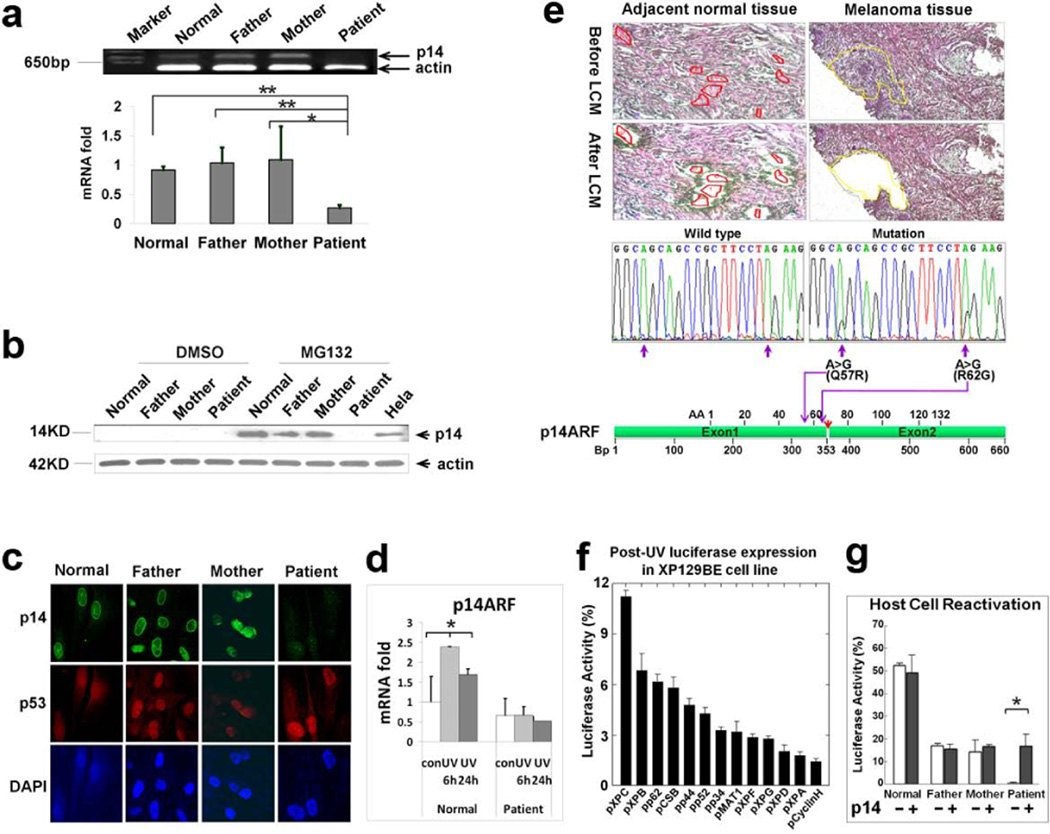
Figure 4.
Mutation, expression and functional analysis of p14ARF. (a) Reduced level of p14ARF mRNA was measured by semi-quantitative RT–PCR (top) and quantitative-real time PCR (bottom) in cells from patient DD129BE, compared to his parents and a normal control. (b) p14ARF protein was measured by western blot. The p14ARF protein level increased markedly in paternal, maternal and normal control fibroblasts in the presence of the proteasome inhibitor (MG132) but p14ARF was not detected in DD129BE cells. (c) Immunofluorescence showed low levels of p14ARF but normal levels of p53 in the patient cells. (d) Relative quantity of p14ARF mRNA in normal and patient fibroblast after UV exposure: cells were irradiated with 10 J/m2 UV and incubated for 6h and 24h. cDNA was synthesized from RNA collected and transcript expression was examined by qRT-PCR. p14ARF mRNA levels did not increase in the patient’s cells at 6 hr or 24 hr after treatment with UV. An asterisk denotes statistically significant difference between control and irradiated normal cells (P < 0.05) while there was no significant difference between control and irradiated patient cells. (e) Identification of p14ARF mutation in melanoma cells isolated by laser capture microdissection (LCM). Adjacent normal and melanoma tissues were microdissected and p14ARF was PCR amplified and sequenced. Two missense mutations were detected in melanoma cells (middle right) but not in adjacent normal tissue (middle left). Locations of two missense mutations are indicated by purple arrows, and the location of the breakpoint in p14ARF is indicated by a red arrow (bottom). (f and g) Post-UV host cell reactivation (HCR). UV-treated or untreated reporter gene plasmid (pCMVLuc) was co-transfected with plasmids expressing wild type cDNA of each of the known XP genes or p14ARF cDNA. (f) Reduced luciferase activity in DD129BE cells was not corrected by co-transfection with known XP genes except for partial correction with wild type XPC cDNA. (g) Co-transfection with wild type p14ARF cDNA-containing plasmid into triplicate cultures of normal (AG13354) (yellow bar), paternal (GM16735) (blue bar), maternal (GM16733) (green bar) and patient (DD129BE (pink bar) fibroblasts. The low luciferase activity in DD129BE cell was increased by co-transfection with wild type p14ARF cDNA. The data represent three independent experiments, and the error bars represent the standard deviation. (a), (d) and (g) mean±s.d., n≥3. *P≤0.05; **P≤0.01.