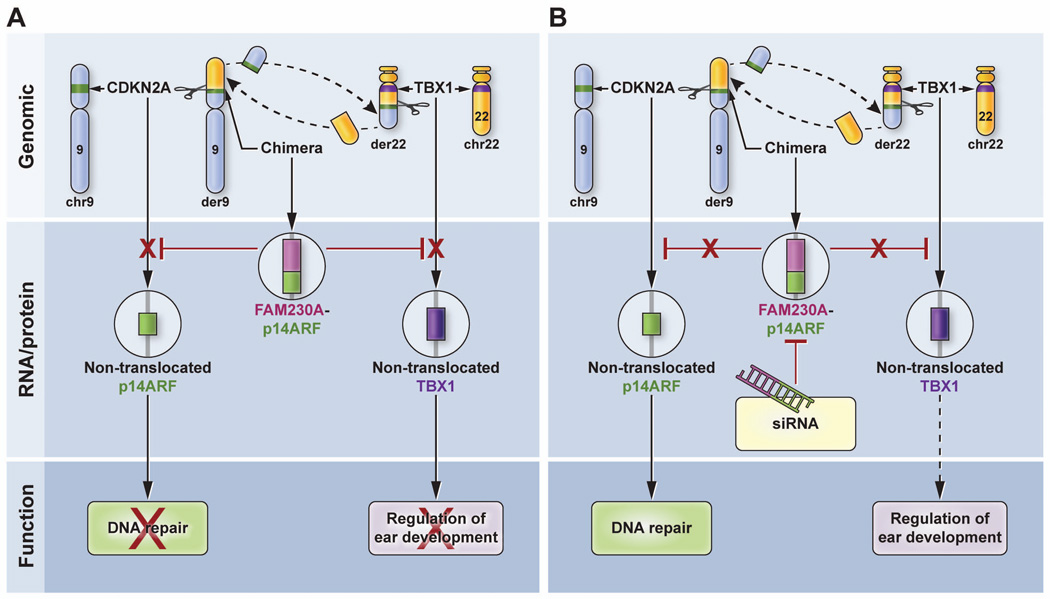
Figure 7.
Model for the action of chimeric p14arf-FAM230A as a dominant negative regulator. A. (Upper panel) The chimeric der9 chromosome is formed by translocation of a portion of the long arm of chromosome 22 (yellow) to the short arm of chromosome 9 (blue) thereby splitting the CDKN2A gene (green band) within p14arf intron 1. The other chromosome 9 is intact with an unaffected CDKN2A gene (green band). The der22 chromosome is formed in a parallel manner. The TBX1 gene (purple band) is located 800kb away from the break and is not involved in the translocation. The other chromosome 22 is unaffected. (Middle panel) Chimeric DGCR mRNA is produced that results in decreased expression of the non-translocated normal p14ARF allele on the other chromosome 9 and decreased expression of both non-mutated TBX1 alleles on chromosome 22. (Lower panel) This dominant negative regulation results in reduced DNA repair and altered fetal development giving rise to DGS-like features. B. Treatment with siRNA (middle panel) results in inhibition of chimeric mRNA which would no longer inhibit expression of p14ARF and TBX1. This would result in increased expression of p14ARF and TBX1. (Lower panel) The increased expression of p14ARF would increase DNA repair and presumably, would facilitate normal ear development.