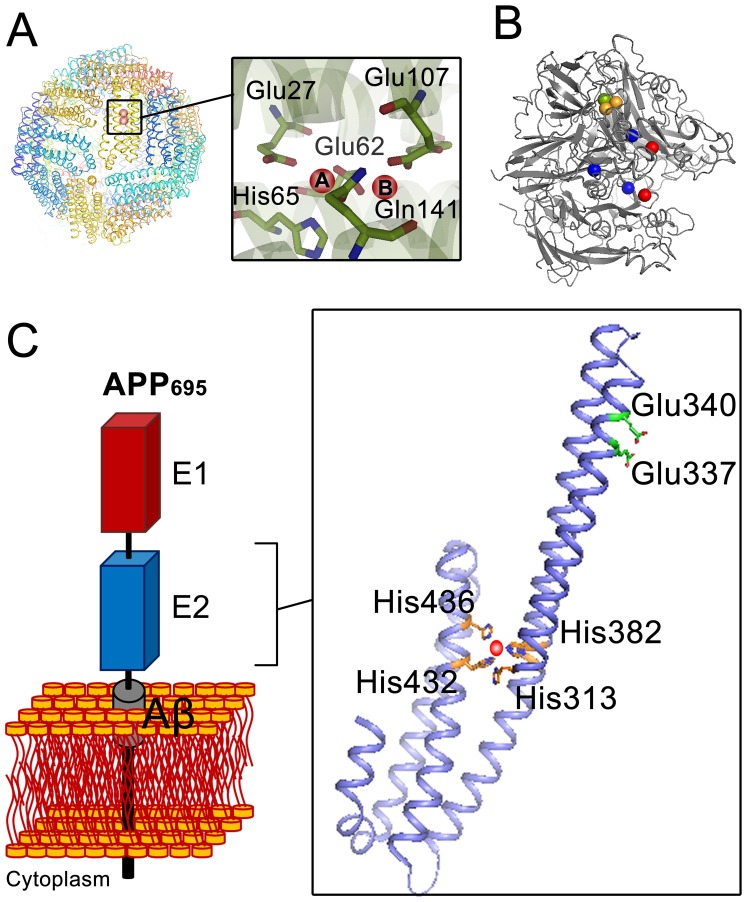
Figure 1. Comparison of the X-ray crystal structure of proteins with ferroxidase activity with that of the E2 domain of amyloid precursor protein (APP).
(A) Quaternary structure of 24-meric ferritin (HuHF, PDB code 2FHA) showing the position of the diiron binding site where the ferroxidase reaction occurs. The two iron binding sites are marked with A and B. (B) Structure of the multicopper oxidase ceruloplasmin (PDB code 1KCW). Ceruloplasmin contains of three type I copper centers (blue sphere), one type II copper center (green sphere), and one type III copper center (orange sphere). Type II and III centers together form a trinuclear copper center which is responsible for four electron oxidation of molecular oxygen to water. Red spheres in the structure show other possible metal binding sites. (C) A schematic representation of the APP and X-ray structure of the E2 domain of APP695 (PDB 3UMH). The structure shows the specific Cu(II) (red sphere) binding site (M1 site) with four histidines (His313, His382, His432, and His436) as coordinating residues. Glu337 and Glu340 are the putative ligands of the previously defined ferroxidase site in the E2 domain of APP [21]. The numbering of the residues is based on APP695.