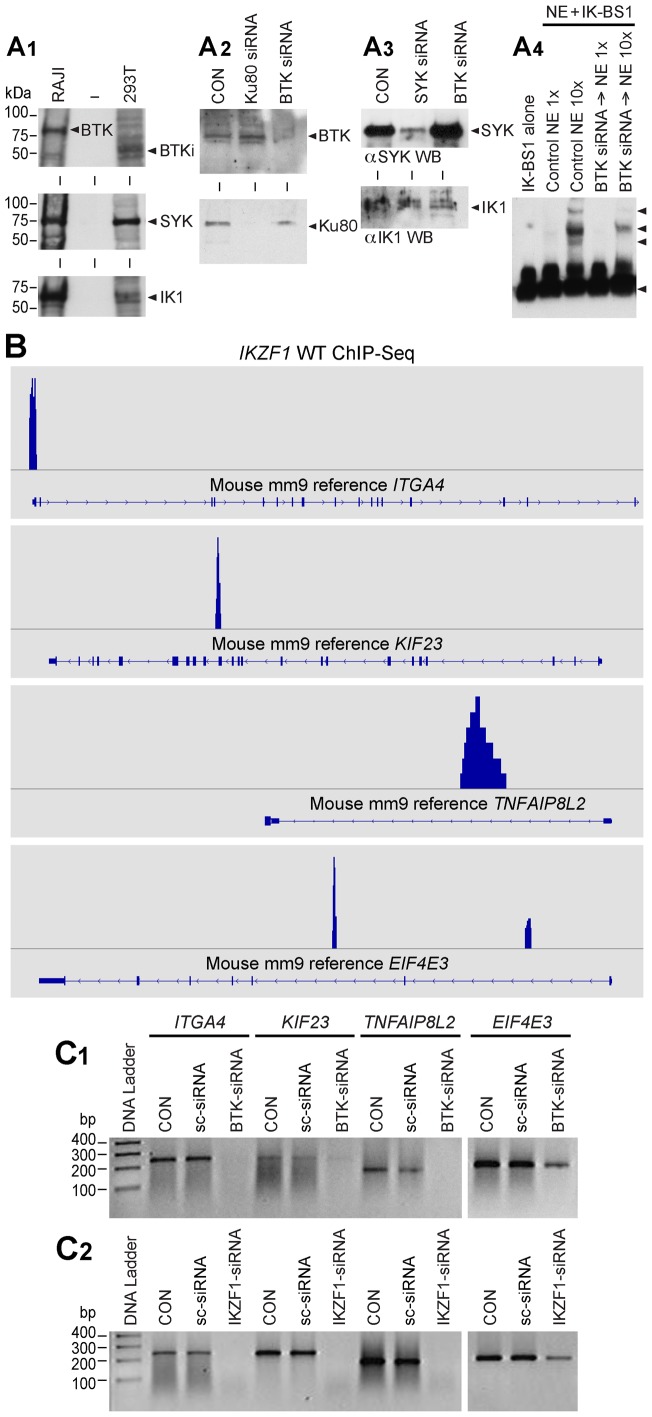
Figure 3. BTK Expression Levels Control DNA Binding Activity and Transcription Factor Function of Ikaros.
[A1] BTK Western Blot Analysis of RAJI and Hek293T cells. RAJI cells expressed predominantly the 77 kDa isoform of BTK, whereas Hek293T cells expressed predominantly a 65 kDa isoform of BTK (labeled as BTKi). Both RAJI and Hek293T cells express SYK and IK1. [A2] Upper panel: BTK Western blot analysis of whole cell lysates from Hek293T cells treated with medium only (CON), BTK siRNA or Ku80 siRNA that was used as a control. Each siRNA was used at a 50 nM concentration. BTK siRNA (but not Ku80 siRNA) resulted in depletion of BTK protein without a decrease in the amount of IK protein. Lower panel : Ku80 Western blot analysis of whole cell lysates from Hek293T cells treated with medium only (CON), BTK siRNA or Ku80 siRNA. BTK siRNA did not affect the expression level of the control protein Ku80. In contrast, Ku80 siRNA resulted in depletion of Ku80 protein. [A3] Additional Controls. SYK vs. IK Western blot analysis of whole cell lysates from 293T cells treated with medium only (CON), SYK siRNA or BTK siRNA [13]. BTK siRNA did not cause a decrease in the amount of SYK or IK proteins [modified from Figure 2 of our recent open access article published in PNAS [13]. [A4] EMSAs were performed on nuclear extracts (NE) from untreated control (CON) Hek293T cells as well as Hek293T cells treated for 72 h with BTK siRNA (50 nM) using biotin-labeled DNA probe IK-BS1. Both 0.4 µg (1×) and 4 µg (10×) amounts of NE were used. IK activity was measured by the electrophoretic mobility shifts of the biotin-labeled IK-BS1 probe, representing IK-containing nuclear complexes (indicated with arrow heads). The position of the probe is also indicated with an arrowhead at the bottom of the gel. The biotin-labeled DNA was detected using a streptavidin-horseradish peroxidase conjugate and a chemiluminescent substrate. The membrane was exposed to X-ray film and developed with a film processor. [B] IK binding sites of validated IK target genes. By cross-referencing IK-regulated gene set (GSE323211) with the archived CHiPseq data (GSM803110) the location of potential IK binding sites for validated IK target genes Itga4 (NM_010576; chr2:79095583–79173271), Eif4e3 (NM_025829; chr6:99575131–99616765), Kif23 (NM_024245; chr9:61765085–61794606) and Tnfaip8l2 (NM_027206; chr3:94943443–94946282) was visualized in the mouse mm9 reference database using the Integrative Genomics Browser. [C] RT-PCR was used to examine the expression levels of randomly selected IK target genes after 72 h treatment with medium alone (CON), 50 nM scrambled siRNA (sc-siRNA), IKZF1 siRNA vs. BTK siRNA. C1: Expression levels of 4 randomly selected IK target genes were reduced by siRNA-mediated depletion of BTK, whereas treatment with sc-siRNA had no such effect. C2: Included as a positive control, IKZF1 siRNA also abrogated or reduced the expression of all 4 IK target genes.