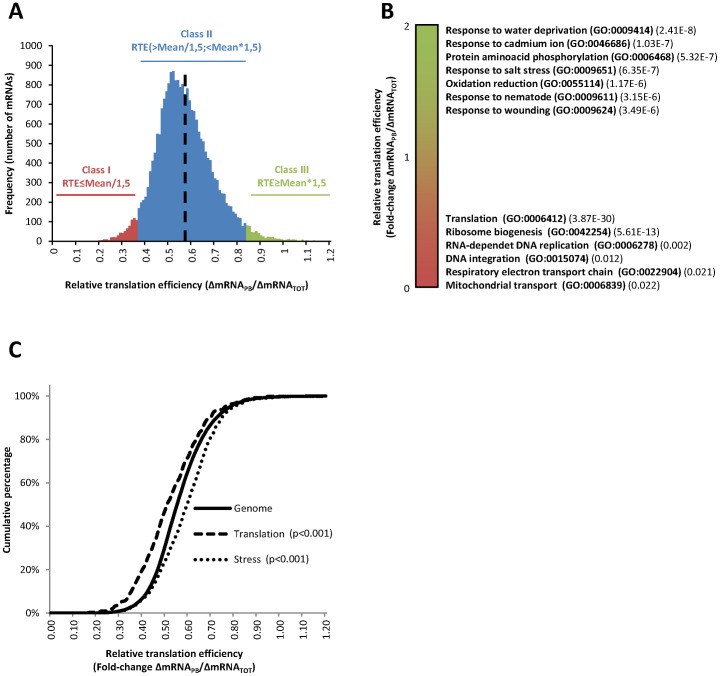
Figure 4. Heat selectively regulates translation of functionally relevant cohorts of mRNAs.
(A) Density distribution of the relative translation efficiencies (ΔmRNAPB/ΔmRNATOT) of the mRNAs selected for the bioinformatics analysis (n = 16098) in response to heat-stress. The average relative translation efficiency of the distribution (0.564) is shown as a dotted black line. The categories, in which mRNAs were classified for subsequent analysis, are also shown in the figure. Class I (red; n = 696) and Class III (green; n = 456) included mRNAs with a relative translation efficiency ≤1.5 fold or ≥1.5 fold the average translation efficiency of the distribution, respectively. Class II (blue; n = 14946) included the rest of the genes within the described parameters. (B) Functional analysis of the selected mRNAs according to their relative translation efficiencies using FatiScan software. Translation efficiencies were graphically ranked in green (ΔmRNAPB/ΔmRNATOT≥1) to red (ΔmRNAPB/ΔmRNATOT≤1) and the most significant GO terms enriched along the ranking were annotated on the right with the adjusted p-values. (C) Distribution (expressed as a cumulative percentage) of the relative translation efficiencies in response to heat stress for mRNAs in the whole Arabidopsis genome (solid line), genes related with the GO term “translation” (dashed line) or genes related with the GO term “stress” (dotted line). K–S test was used to statistically examine the variations in the distributions, using the whole genome as reference. The corresponding p-values are indicated in the figure.