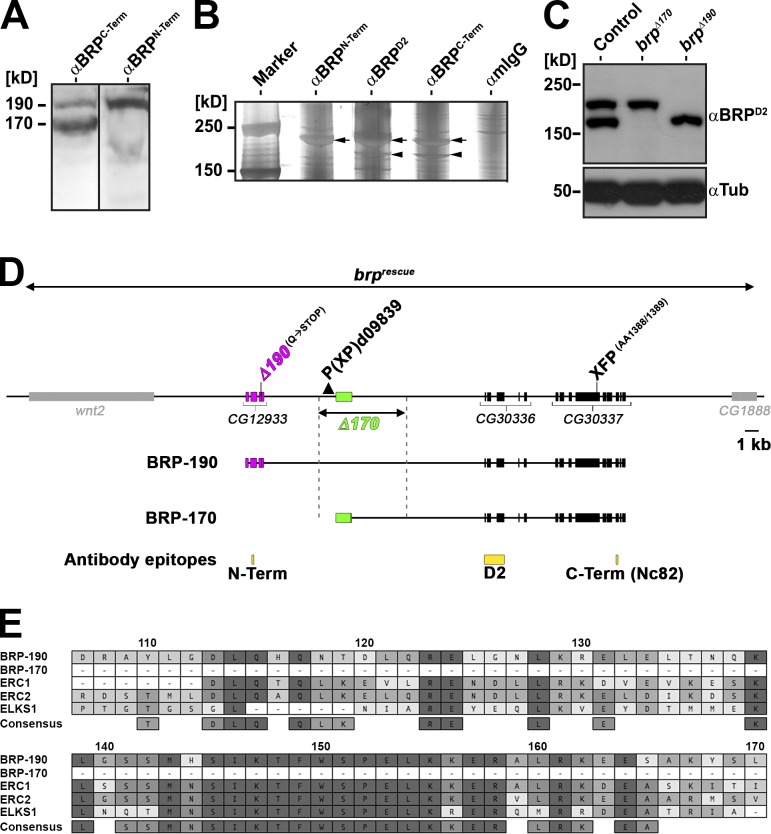
Figure 1.
The brp locus encodes two major isoforms. (A) Western blots of Drosophila adult head extracts probed with anti-BRPC-Term AB (short AB) and anti-BRPN-Term AB. Black line indicates that intervening lanes have been spliced out. (B) Silver gel of IPs conducted on Drosophila adult head extracts with the indicated ABs. Arrows represent the two BRP isoforms (top arrows: BRP-190; bottom arrows: BRP-170). (C) Western blot of Drosophila adult head extracts of the indicated genotypes probed with anti-BRPD2 and anti-Tubulin (Tub). (D) Schematic representation of brp genomic locus. BRP-190 and BRP-170 have distinct N-terminal sequences (magenta, BRP-190; green, BRP-170) but share the same C-terminal exons (black). AB epitopes are indicated in yellow. brpΔ190 allele: ethyl methyl sulfonate–induced point mutation from CAG (Glutamine [Q]) to TAG, leading to a premature STOP at aa 261 (encoded by the BRP-190–exclusive exon 3). brpΔ170 was generated by imprecise excision of P element P(XP)d09839 deleting the region indicated by the dashed lines. Genomic rescue construct (brprescue) encompassing the whole locus including two neighboring genes. (E) Protein sequence alignment (MAFFT with BLOSUM62 scoring matrix) between Drosophila BRP-190 and BRP-170, mouse ERC1 (NCBI Nucleotide Database accession no. NP_835186) and ERC2 (NCBI Nucleotide Database accession no. NP_808482), and Caenorhabditis elegans ELKS1 (NCBI Nucleotide Database accession no. NP_500329). Numbering is based on BRP-190. Similarity in amino acid identity is indicated by a color code (dark gray, 100%; gray, 80–100%; light gray, 60–80%; white, <60% similarity). Term, terminal.