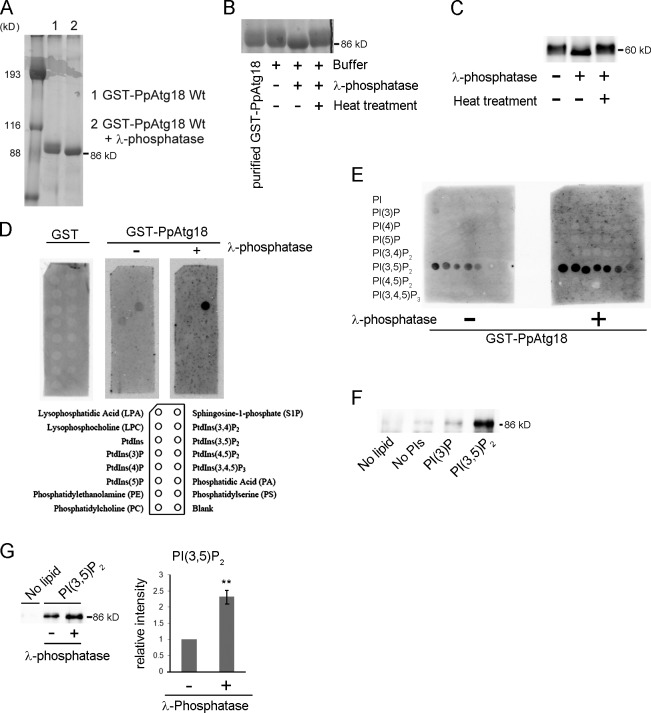
Figure 1.
Phosphorylation of PpAtg18 modulates PI(3,5)P2-binding activity. (A) Evaluation of purified GST-PpAtg18 from P. pastoris by SDS-PAGE and Coomassie brilliant blue staining. Lane 1: Purified GST-PpAtg18 was eluted from a GS 4B column using the reduced form of glutathione. Lane 2: Purified GST-PpAtg18 was treated with λ-phosphatase on the GS 4B column. This sample was used for the lipid binding assay. (B) Phosphatase treatment of purified PpAtg18 from P. pastoris. Heat, λ-phosphatase was inactivated at 65°C for 1 h. (C) Phosphatase treatment of cell-free extracts from the strain expressing PpAtg18-5×Flag under the original PpATG18 promoter. The cell lysate was treated with λ-phosphatase at 37°C for 1 h. (D) PIP strip analysis. Membranes were incubated with 0.5 µg/ml protein and detected using the Light Capture II system (ATTO). GST-PpAtg18Wt and GST-Atg18Wt with λ-phosphatase were acquired simultaneously to ensure equal exposure times. (E) PIP array analysis. Membranes were incubated with 1.0 µg/ml protein and detected simultaneously to ensure equal exposure times. (F) Liposome pull-down assay. Purified GST-PpAtg18 (1.0 µg) was incubated with each liposome preparation and centrifuged at 16,000 g for 20 min. The pellets were washed twice, suspended in sample buffer, and analyzed by immunoblotting to detect GST-PpAtg18. No lipid, no liposomes; No PI, liposomes without PIs; PI(3)P, liposome containing PI(3)P; PI(3,5)P2, liposome containing PI(3,5)P2. (G) Liposome pull-down with dephosphorylated PpAtg18. Each protein (1.0 µg) was incubated with liposomes containing PI(3,5)P2 and analyzed by immunoblot to detect GST-PpAtg18. The bands were analyzed by densitometry, and band intensities were normalized to samples not treated with phosphatase. Error bars indicate mean ± SEM. **, P < 0.05.