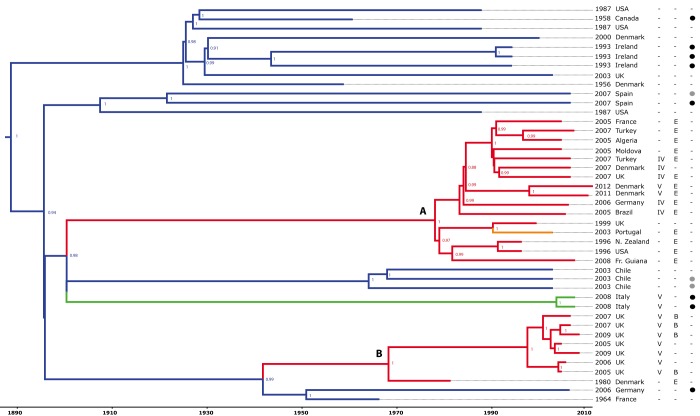
FIG 1 .
Identification of human epidemic S. aureus clones descended from bacteria that made livestock-to-human host jumps. The Bayesian phylogenetic reconstruction of the CC97 lineage is shown. The tree is based on core genome alignment with branches color coded according to the host species association (blue, bovine; green, porcine; orange, caprine; red, human) and date and country of origin of each isolate indicated. The presence or absence (−) of β-toxin phage IEC variants B and E and SCCmec type IV or V is indicated by the appropriate letter, and the presence of S. aureus pathogenicity island (SaPI)-encoded vwb or phage-encoded lukM/lukF is denoted by black and gray circles, respectively. The branch lengths are scaled according to the time scale bar (years) and the posterior probability values are indicated at each node. Clades A and B are shown.