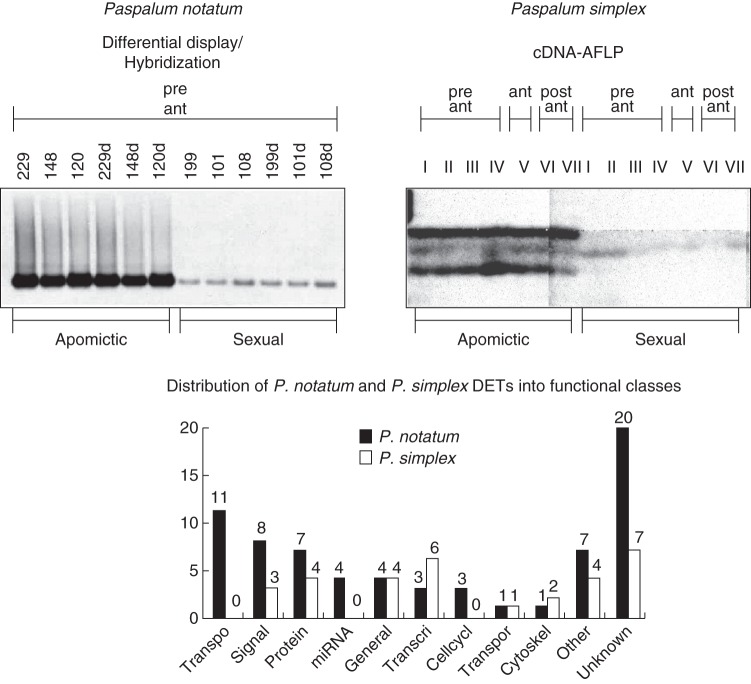
Fig. 5.
Characterization of apomixis-associated DETs identified by transcriptomic analysis in Paspalum genotypes. Top left: a representative example of DET (apo417/N114) identified in P. notatum flowers at late pre-meiotic/meiotic stage by differential display experiments. Differential display amplicons originating from three apomictic and three sexual F1 plants (in duplicate) were transferred to nylon membranes and hybridized with an N114 probe labelled with digoxigenin (Pessino et al., 2001). Top right: a representative example of a Class A DET identified in P. simplex flowers by cDNA-AFLP analysis at pre-anthesis, anthesis and post-anthesis stages (Polegri et al., 2010). Bottom: histogram representing the relative occurrence of DETs belonging to particular ontology classes in P. notatum, as identified from DD experiments and in P. simplex, as identified from cDNA-AFLP experiments. Ontology classes (from left to right): transposon/retrotransposon, signal transduction, protein metabolism, miRNA precursors, general metabolism, transcription, cell cycle, transport, cytoskeleton, other, unknown.