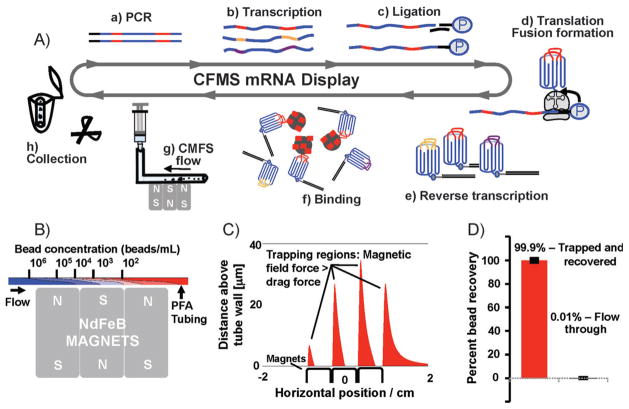
Figure 1.
CFMS mRNA display selection. A) In vitro selection of mRNA-display libraries using CFMS. 10Fn3 scaffold: blue; random regions: red, gold, and purple. The 10Fn3 scaffold is represented as greek key motif with random BC and FG loops. Selections begin with PCR amplification (a) to produce copies of approximately one trillion unique sequences, in vitro run-off transcription (b), splint-mediated ligation of a 3′ puromycin-containing oligonucleotide (c), in vitro translation and fusion formation (d), and subsequent reverse transcription (e), pool binding (f), and affinity enrichment. Target-coated beads are captured within the PFA tubing by three small NdFeB magnets (g) and washed under continuous flow. Bead recovery is achieved by cutting a minimal length of tubing containing the beads and eluting directly into the PCR mix (h). B) Simulation of the depletion in bead concentration resulting from vertical magnetic capture. C) Simulation showing the regions in the tube where the magnetic field is sufficient to retain beads attracted to the tube surface against the fluid flow. In the regions shown in red, the magnetic force in the negative horizontal direction exceeds the maximum Stokes drag force experienced by a stationary bead in the flow field within the tube. We identified four regions that extend to a distance of 10–30 microns into the tubing. D) Bead recovery analysis using FACS. All but 0.01% of beads applied to the device were successfully trapped.