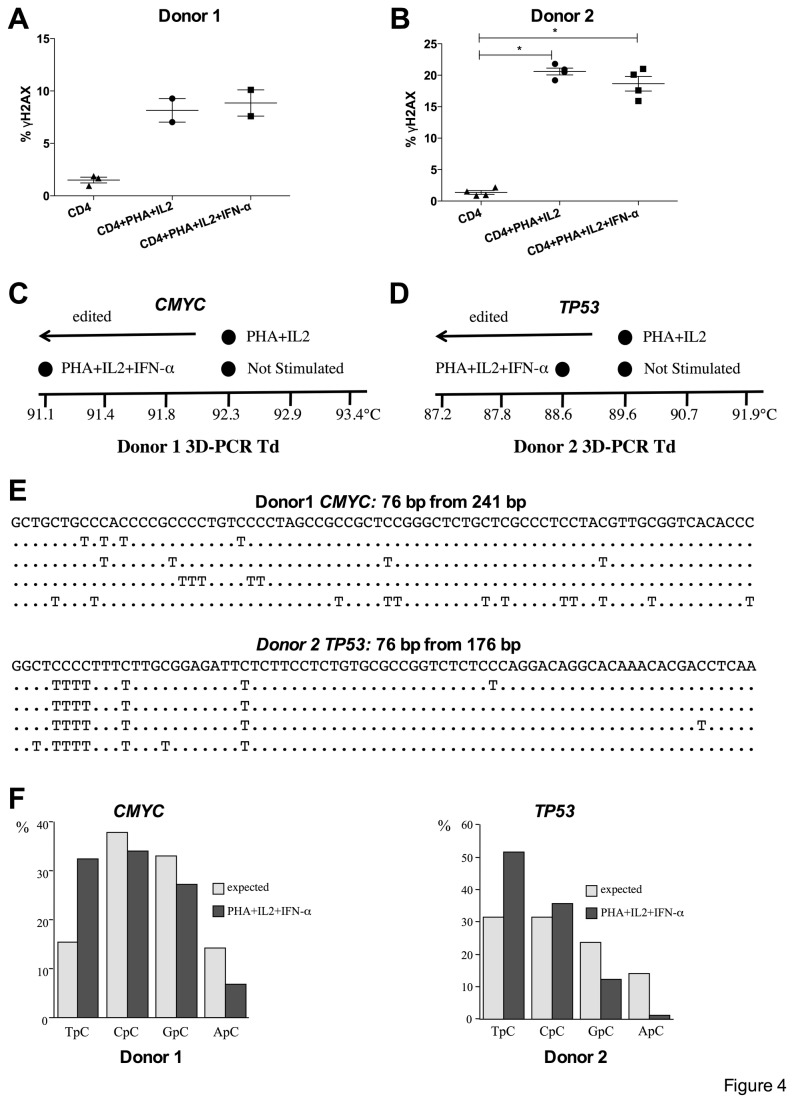
Figure 4. Induction of DSBs and A3A editing in activated primary human CD4+ T lymphocytes.
(A) (B) ©H2AX positive DSBs in activated CD4+ T lymphocytes. Mean and SEM are shown. Group comparisons were calculated using the Mann-Whitney test (*p < 0.05). (C) (D) CD4+ T lymphocytes were transduced by recombinant lentivirus encoding the UNG inhibitor UGI (rV2.EF1.UGI). Recovery of hyperedited CMYC DNA by 3DPCR from donor 1 (C) and TP53 DNA from donor 2 as shown by the denaturation temperature (Td) of the 3DPCR products (D). Only for the PHA+IL2+IFN-α treatment APOBEC3 edited DNA was recovered. The difference in minimal denaturation temperatures is due to the different base composition of the CMYC and TP53 fragments. (E) A selection of hyperedited CMYC (Donor 1) or TP53 (Donor 2) sequences respectively are shown compared to the unedited sequence. Only differences are shown. For space reasons only a fraction of the sequences are shown. (F) 5’ dinucleotide context associated with editing along with expected values assuming no editing bias. The clear preference for TpC is a diagnostic trait of A3A editing of nuDNA.