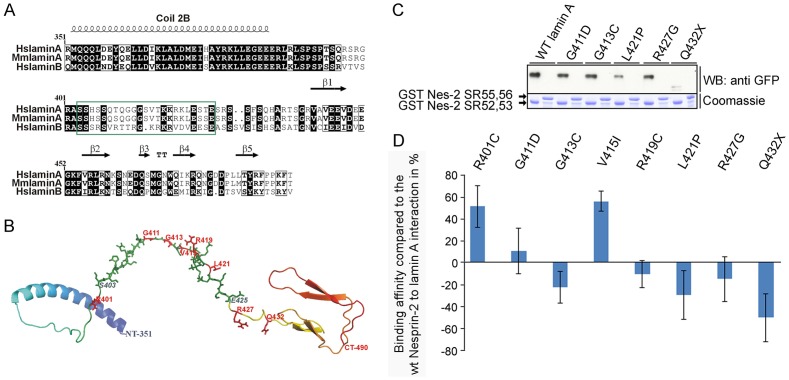
Figure 4. The interaction site of lamin A to Nesprin-2 is in a loop and mutations in LMNA modulate the interaction to Nesprin-2.
(A) Amino acid alignment of a human lamin A/C amino acids 351–490 (HsLaminA; uniprot accession number P02545) and the corresponding sequence in human lamin B (HsLamin; P20700) and Mus musculus lamin A (MmLaminA; P48678). Secondary structure elements are shown on top of the alignment and the lamin A region that interacts with Nesprin-2 is boxed in green. Conserved residues are highlighted black, similar residues are boxed. (B) Prediction of the three dimensional structure of human lamin A aa 351–490. The three dimensional structure of human Lamin A351–490 was predicted by using multiple PDB structures as templates (1UFGA, 1IFRA, and 2LLA) in the MULTICOM server [41]. The lamin A interaction site aa 403–425 to Nesprin-2 is highlighted in green and aa 403 and 425 are pointed out in blue. Amino acids that targets for the mutations analysed here are highlighted in red. The structure prediction was generated by using pyMOL v1.3. (C) The binding properties between WT GST Nesprin-2 SR52,53 and GFP lamin A mutations were analysed by pull down experiments. COS7 cells expressing WT or mutated GFP lamin A proteins were lysed and incubated with recombinant GST Nesprin-2 SR52,53 proteins. GST Nesprin-2 SR55,56 proteins were used as negative controls. (D) WT and mutant GFP lamin A proteins show distinct binding properties to WT GST Nesprin-2 SR52,53. The zero baseline represents the 100% binding affinity between WT GST Nesprin-2 SR52,53 and GFP lamin A (for details see materials and methods). Deviations caused by distinct mutations in LMNA are given in percent. Each mutation was analysed by four to seven independent experiments.