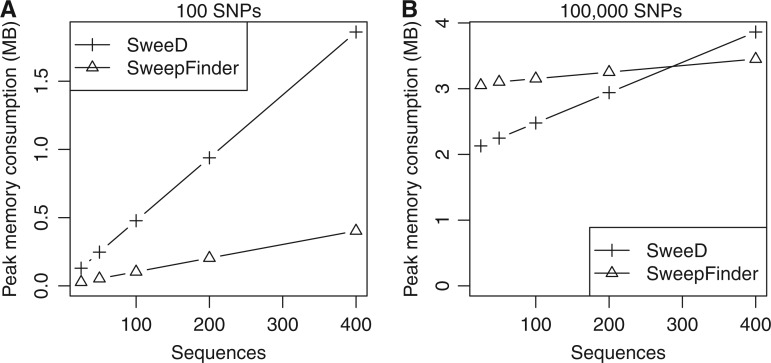
Fig. 1.
Comparison of peak memory consumption between SweeD and SweepFinder. Simulated data sets of 100 SNPs (A) and 100,000 SNPs (B) and 25, 50, 100, 200, and 400 respective sequences were used for the measurements. Memory consumption was quantified with the massif tool of the valgrind software (Seward and Nethercote 2005). In most cases, SweeD consumes more memory than SweepFinder due to the lookup table implementation. However, memory consumption is in the order of MBs even for very large data sets.