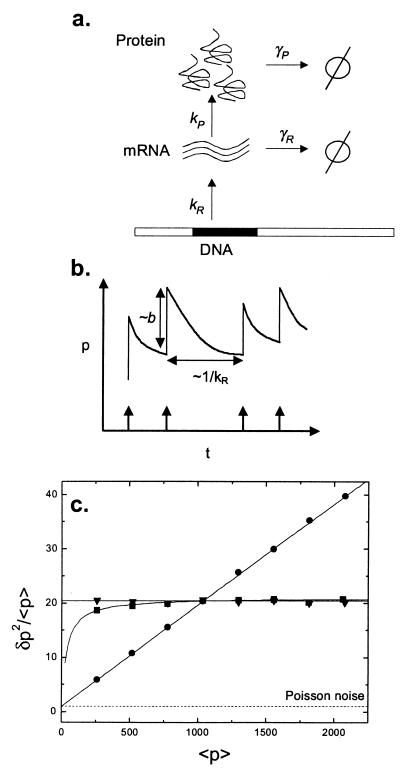
Figure 1.
Modeling single gene expression. (a) mRNA molecules are synthesized at rate kR from the template DNA strand. Proteins are translated at a rate kP off each mRNA molecule. Proteins and mRNA degrade at rates γP and γR, respectively. Degradation into constituents is denoted by φ. All reactions are assumed to be Poisson, so that the probability of a reaction with rate k happening in a time dt is given by kdt, and the waiting times between successive reactions are exponentially distributed. (b) A simplified timecourse illustrates the intuition behind the result of Eq. 1. Transcription initiation events occur with average frequency kR and are indicated by arrows; each mRNA transcript releases a burst of proteins of average size b, and proteins decay between bursts. (c) Controlling mean and variance of protein number by varying system parameters for single gene expression. The Fano factor ratio (δp2/<p> ≡ variance/mean) is plotted versus the mean. The mRNA half-life is fixed at 2 min. The base case corresponds to a burst size b = 20, a transcript initiation rate kR = 0.01 s−1 and a protein half-life ln(2)/γP = 1 h. The three curves are produced by varying one of these parameters while keeping the other two fixed. b is varied between 5 and 40 (circles); kR is varied between 0.0025 s−1 and 0.02 s−1 (triangles); protein half-life is varied from 15 min to 2 h (squares). The Poisson value of δp2/<p> = 1 is marked for comparison. Monte Carlo results (symbols) match analytic values given by Eq. 1 exactly (solid lines).