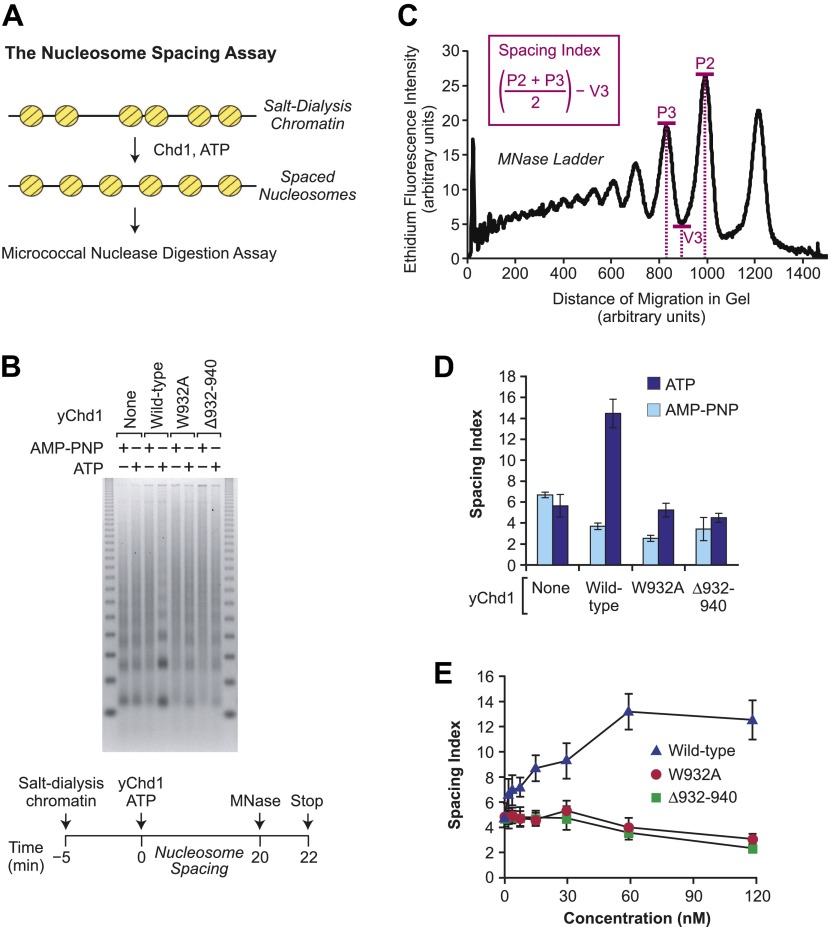
Figure 4. The chromatin remodeling-defective yChd1 proteins are not able to catalyze the formation of regularly-spaced nucleosomes.
(A) Schematic representation of the nucleosome spacing reaction. Randomly-distributed nucleosomes were generated by salt dialysis reconstitution of chromatin. Wild-type or mutant Chd1 protein was added along with ATP, and the reaction products were characterized by partial MNase digestion analysis. (B) Mutant yChd1 proteins do not reposition nucleosomes into periodic arrays. Spacing reactions were performed by incubating salt-dialysis chromatin with wild-type or mutant yChd1 proteins (30 nM) in the presence of either AMP-PNP or ATP. The reaction products were characterized by partial MNase digestion analysis. (C) Determination of the spacing index. Agarose gels from nucleosome spacing reactions were stained by ethidium bromide and then subjected to imaging and analysis on ImageQuantTL (GE) to obtain densiometry scans. The spacing index is the average height of the di- and tri-nucleosome peaks ([P2 + P3)/2) minus the height of the valley (V3) between the peaks, as indicated by the formula shown in the figure. (D) Quantitative analysis of nucleosome spacing by wild-type and mutant yChd1 proteins. The spacing indices were determined for the products of reactions such as those shown in (B). The results are presented as the mean ± standard deviation (N = 6). (E) The chromatin remodeling-defective yChd1 proteins exhibit little to no activity in the nucleosome spacing assay at different concentrations. Nucleosome spacing reactions were performed with yChd1 proteins and subjected to partial MNase digestion analysis. The graph depicts the average spacing index ± standard deviation (N = 4) vs concentration (nM) for each of the indicated proteins.