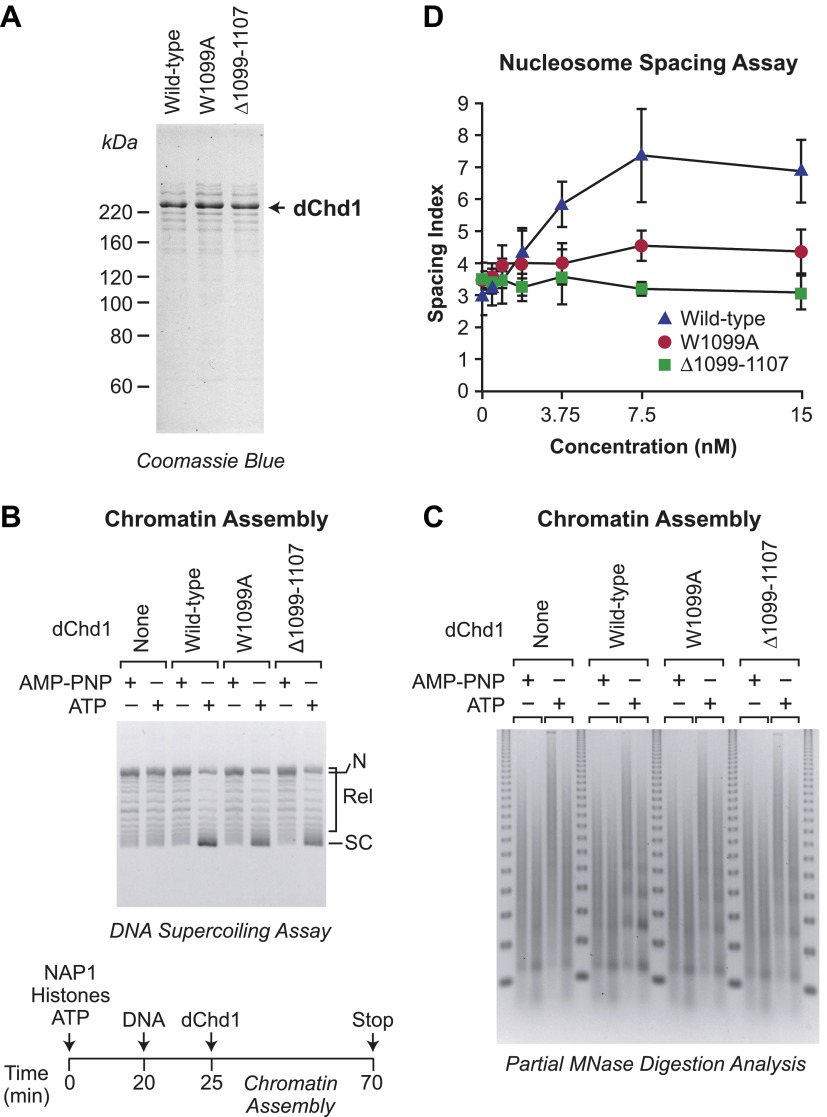
Figure 5. ATP-dependent nucleosome assembly is observed with chromatin remodeling-defective Drosophila Chd1 (dChd1) proteins.
(A) Purification of recombinant wild-type and mutant dChd1. The purified proteins were analyzed by polyacrylamide-SDS gel electrophoresis and visualized by staining with Coomassie Blue. (B) Chromatin remodeling-defective dChd1 proteins can assemble nucleosomes in an ATP-dependent manner. Chromatin assembly reactions were performed with wild-type or mutant dChd1 proteins (60 nM) in the presence of either AMP-PNP or ATP. The reaction products were analyzed by the DNA supercoiling assay. The positions of supercoiled (SC), relaxed (Rel), and nicked open circular (N) DNAs are indicated. (C) Chromatin assembly with remodeling-defective dChd1 proteins does not yield regularly-spaced nucleosomes. Chromatin assembly reactions were performed with wild-type or mutant dChd1 proteins (60 nM) in the presence of either AMP-PNP or ATP. Reaction products were subjected to partial MNase digestion analysis. (D) The chromatin remodeling-defective dChd1 proteins exhibit reduced nucleosome spacing activity. Spacing assays were performed with dChd1 proteins and subjected to partial MNase digestion analysis. This graph depicts the mean spacing index ± standard deviation (N = 3) vs concentration (nM).