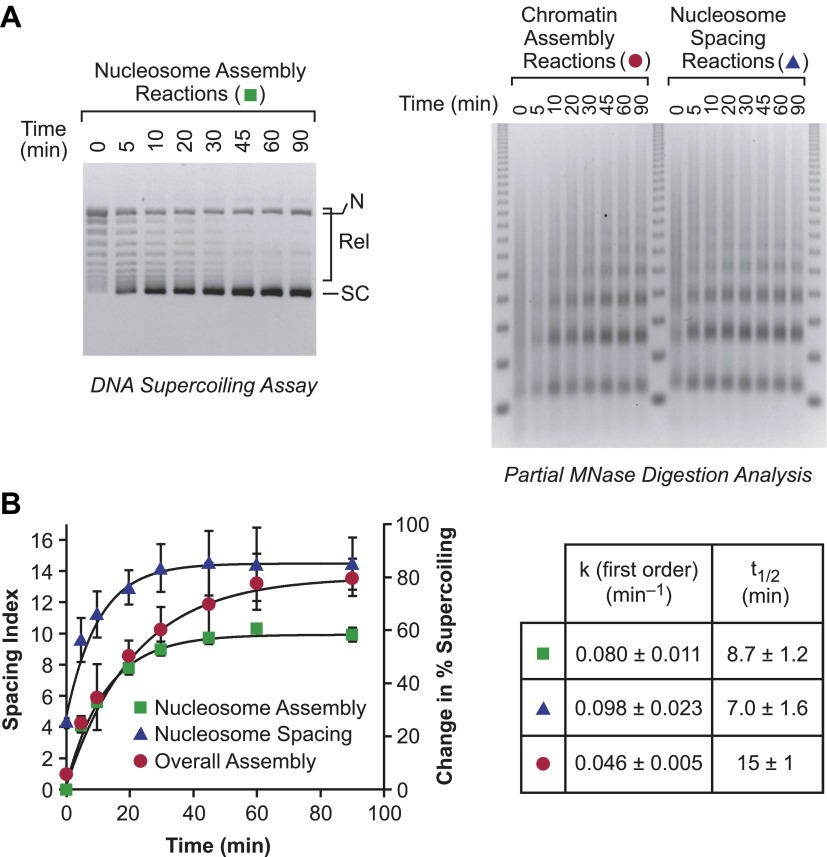
Figure 8. The individual rates of nucleosome formation and spacing are faster than the overall rate of assembly of periodic arrays of nucleosomes.
(A) Determination of the rates of nucleosome formation, nucleosome spacing, and assembly of regularly-spaced nucleosomes. Nucleosome formation in chromatin assembly reactions was monitored by using the DNA supercoiling assay. The positions of supercoiled (SC), relaxed (Rel), and nicked open circular (N) DNAs are indicated. Nucleosome spacing reactions were analyzed by partial MNase digestion analysis. The overall assembly of periodic arrays of nucleosomes was determined by performing chromatin assembly reactions and analyzing the reaction products by partial MNase digestion analysis. All reactions were performed with wild-type yChd1 at 30 nM. (B) Quantitation of the nucleosome formation and spacing assays, such as those shown in (A). The figure displays the change in % supercoiling ([Δ supercoiled DNA/total DNA] × 100%) and spacing indices vs reaction time (min). The reactions followed first-order kinetics, and the first-order rate constants (min−1) and half-times for reaction (t1/2, min) are given in the table. The data points are presented as mean ± standard deviation (N = 3), and are depicted with the plots of the first order curves (r2 > 0.95 for all graphs).