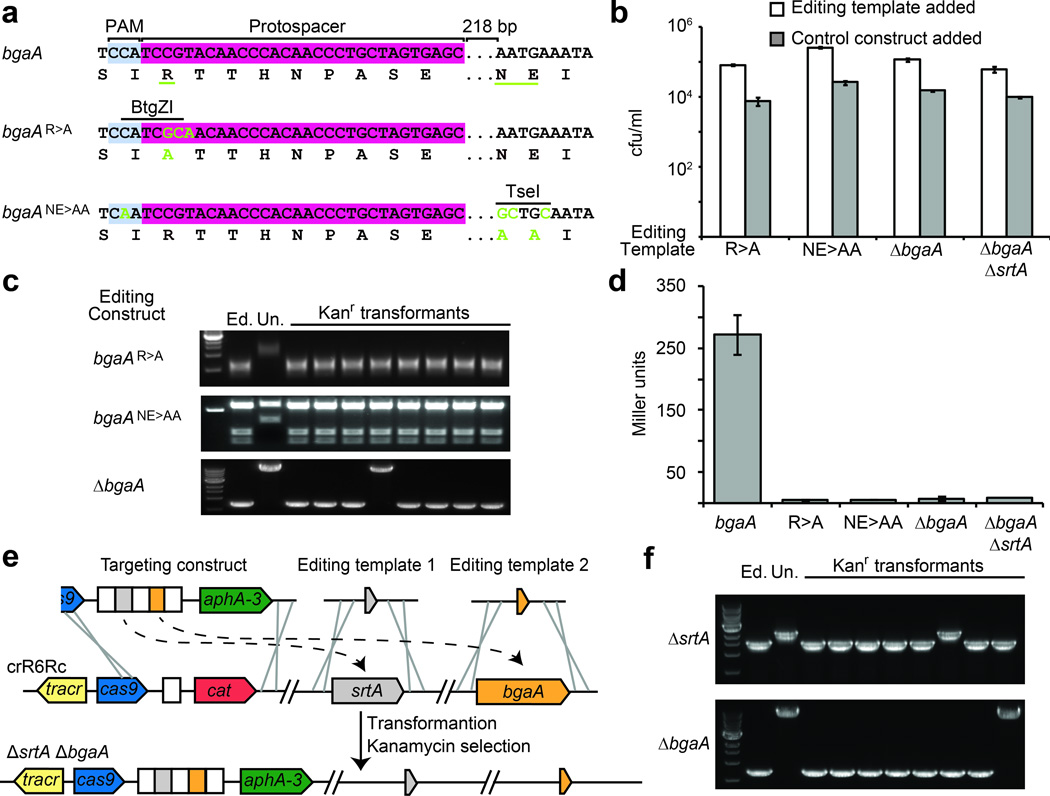
Figure 3.
Introduction of single and multiple mutations using CRISPR-mediated genome editing. (a) Nucleotide and amino acid sequences of the wild-type and edited (green nucleotides; underlined amino acid residues) bgaA. The protospacer, PAM and restriction sites are shown. (b) Transformation efficiency of cells transformed with targeting constructs in the presence of an editing template or control. (c) PCR analysis for 8 transformants of each editing experiment followed by digestion with BtgZI (R>A) and TseI (NE>AA). Deletion of bgaA is revealed as a smaller PCR product. (d) Miller assay to measure the β-galactosidase activity of WT and edited strains. (e) For a single-step double deletion the targeting construct contains two spacers (in this case matching srtA and bgaA) and is co-transformed with two different editing templates (f) PCR analysis for 8 transformants to detect deletions in srtA and bgaA loci. 6/8 transformants contained deletions of both genes.