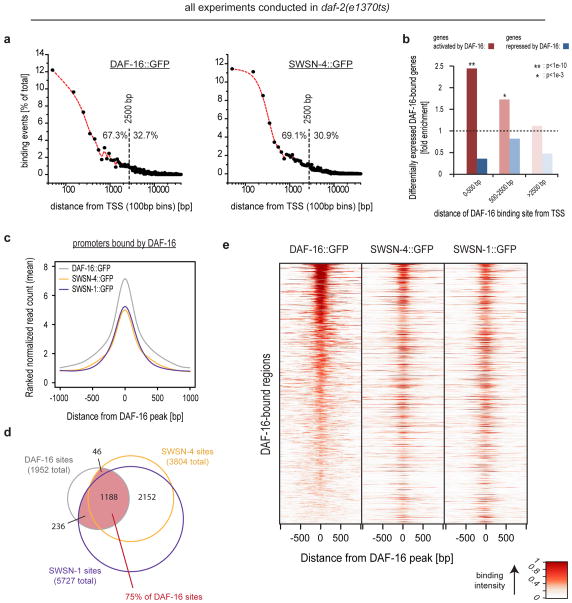
Figure 4.
SWI/SNF extensively associates with DAF-16/FOXO-bound promoter regions. C. elegans of either daf-2(e1370ts); DAF-16::GFP, daf-2(e1370ts); SWSN-4::GFP, or daf-2(e1370ts); SWSN-1::GFP were grown asynchronously and e1370ts was inactivated by a 20h shift to restrictive temperature. ChIP-Seq of GFP-tagged proteins was performed and binding sites were determined. (a) Each binding site was associated with its closest transcriptional start site (TSS) and distances were plotted, revealing that DAF-16/FOXO and SWI/SNF are predominantly located within 2.5 kb of a TSS and thus within promoter regions. (b) Genes in vicinity to a DAF-16/FOXO binding site are strongly enriched for DAF-16/FOXO-activated and rather depleted for DAF-16/FOXO-repressed genes. This effect decays with the distance of the binding site from the TSS. (c) DAF-16/FOXO bound promoter regions are strongly enriched for binding by SWSN-4::GFP and SWSN-1::GFP. Mean read distributions across all DAF-16/FOXO binding sites are shown for the indicated strains. (d) Venn diagram showing the overlap between binding sites of DAF-16/FOXO, SWSN-4/BRG1, and SWSN-1/BAF155/170. (e) Heat-map representation of the data contributing to (c,d), further supporting that SWI/SNF binding occurs at the majority of DAF-16/FOXO binding sites. Lines of the heat-map represent the individual DAF-16/FOXO-bound regions and are sorted by intensity of DAF-16/FOXO binding.