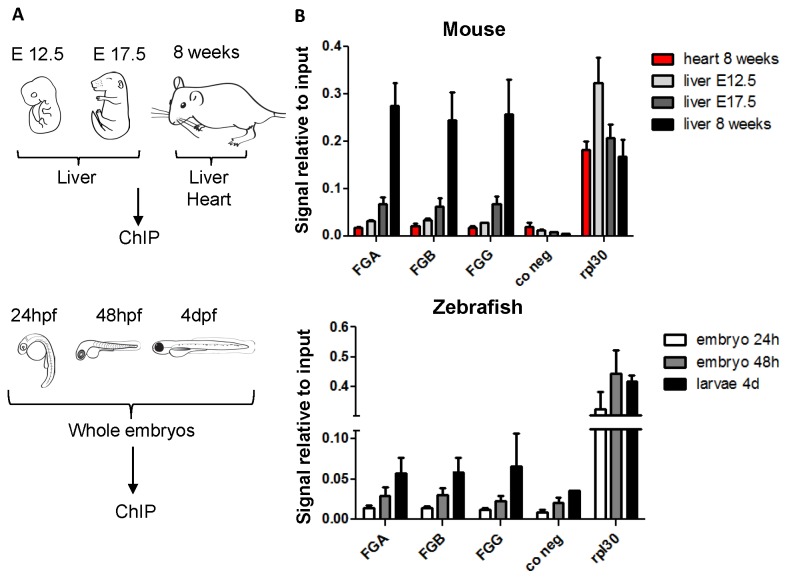
Figure 4. Active chromatin mark H3K4me3 on the fibrinogen promoters during mouse and zebrafish development.
(A) Overview of the samples used for Chromatin immunoprecipitation (ChIP). For mouse experiments, the 8 week tissues are the same as those used for qPCR and DNA methylation sequencing, the E17.5 and E12.5 mice are from the same crossing as the embryos used for Figure 3. For zebrafish experiments, ChIP was performed on whole embryos at different stages. All experiments were performed with three biological replicates, except for mouse E12.5 liver, where two pools of five livers were used. (B) Enrichment of H3K4me3 on mouse and zebrafish fibrinogen promoters and control regions. After ChIP on tissues or whole zebrafish embryos the signal relative to the total input was determined by qPCR, using primers situated around the transcription start site or in the first intron of each fibrinogen gene (Table S1). Primers binding to the first intron of the ubiquitously expressed Rpl30 gene serve as a positive control for enrichment. As a negative control for the H3K4me3 enrichment, gene free regions on chromosome 12 (mouse) and 14 (zebrafish) were chosen. The mean enrichment plus SEM is shown. Anti-rabbit IgG was used as a negative control in the ChIP experiments and the signals per total input were <0.0007 for all genes examined.