Abstract
DNA mismatch repair during replication is a conserved process essential for maintaining genomic stability. Mismatch repair is also implicated in cell-cycle arrest and apoptosis after DNA damage. Because yeast and human mismatch repair systems are well conserved, we have employed the budding yeast Saccharomyces cerevisiae to understand the regulation and function of the mismatch repair gene MSH2. Using a luciferase-based transcriptional reporter, we defined a 218-bp region upstream of MSH2 that contains cell-cycle and DNA damage responsive elements. The 5′ end of the MSH2 transcript was mapped by primer extension and was found to encode a small upstream open reading frame (uORF). Mutagenesis of the uORF start codon or of the uORF stop codon, which creates a continuous reading frame with MSH2, increased Msh2 steady-state protein levels ~2-fold. Furthermore, we found that the cell-cycle transcription factors Swi6, Swi4, and Mbp1—along with SCB/MCB cell-cycle binding sites upstream of MSH2—are all required for full basal expression of MSH2. Mutagenesis of the cell-cycle boxes resulted in a minor reduction in basal Msh2 levels and a 3-fold defect in mismatch repair. Disruption of the cell-cycle boxes also affected growth in a DNA polymerase-defective strain background where mismatch repair is essential, particularly in the presence of the DNA damaging agent methyl methane sulfonate (MMS). Promoter replacements conferring constitutive expression of MSH2 revealed that the transcriptional induction in response to MMS is required to maintain induced levels of Msh2. Turnover experiments confirmed an elevated rate of degradation in the presence of MMS. Taken together, the data show that the DNA damage regulation of Msh2 occurs at the transcriptional and post-transcriptional levels. The transcriptional and translational control elements identified are conserved in mammalian cells, underscoring the use of yeast as a model system to examine the regulation of MSH2.
Keywords: Mismatch repair, Cell-cycle, DNA damage, Transcription, Promoter, Yeast
1. Introduction
Colorectal cancer is the third leading cause of cancer death in the United States [1]. An estimated 10–20% of colorectal cancer cases are attributable to an inherited susceptibility [reviewed in 2]. The most common form of inherited colorectal cancer is hereditary non-polyposis colorectal cancer or Lynch Syndrome, an autosomal dominant disorder characterized by a paucity of colonic polyps, a penetrance of nearly 90%, an early age of onset, and an elevated risk of extracolonic cancer, including cancers of the stomach, endometrium, and small intestine [reviewed in 2]. These tumors display genome-wide microsatellite instability [3–5]. This distinctive instability, reminiscent of the phenotype of DNA mismatch repair defective bacteria [6] and yeast [7], played a crucial role in linking this cancer syndrome to defects in mismatch repair [8–10].
Postreplicative DNA mismatch repair is essential for maintaining genome stability [reviewed in 11]. By eliminating single-base mismatches and insertion-deletion loops that arise due to DNA polymerase error and slippage, mismatch repair enhances the fidelity of DNA replication 1000-fold [reviewed in 12]. In addition to its role in repair of spontaneous mutations, mismatch repair is crucial for induction of appropriate cellular responses to DNA damage [reviewed in 13].
Mutant Escherichia coli with elevated spontaneous mutation rates allowed for the earliest characterization of DNA mismatch repair and further research determined that the mismatch repair proteins are conserved from bacteria to mammals [reviewed in 11]. The basic events of DNA mismatch repair include recognition and binding of a mismatch by MutS homologs (Msh proteins), followed by subsequent events mediated by MutL homologs (Mlh proteins) [reviewed in 14]. Important sequential events include cleavage and degradation of the error-containing strand followed by re-synthesis by DNA replication components [reviewed in 12].
Because of the striking similarity between yeast and human mismatch repair, we employ the yeast Saccharomyces cerevisiae to examine aspects of mismatch repair regulation. MutS Homolog 2 (MSH2) is essential for mismatch recognition and displays cell-cycle periodicity of expression in yeast [15,16]. Specifically, yeast MSH2 transcripts peak at the G1/DNA synthesis boundary (G1/S). Additionally, yeast MSH2 mRNA levels are increased upon treatment with DNA damaging agents, including methyl methanesulfonate (MMS) [17], hydroxyurea (HU) [18], and camptothecin [19].
Human MSH2 mRNA levels are up regulated by E2F expression, consistent with cell-cycle regulation [20–22]. Investigators have shown that human MSH2 mRNA and protein levels change during the cell cycle [23], while others dispute the cell cycle regulation [24]; however, the apparent difference may be due the way in which the cells were prepared for synchrony [23]. Finally, mouse MSH2 has been shown to be cell cycle regulated [25]. Taken together, there is mounting evidence that mammalian MSH2 is cell-cycle regulated.
Silencing of the promoter region of the MLH1 DNA mismatch repair gene has been linked to mismatch repair dysfunction and cancer progression [26]. The profound clinical consequences of promoter dysfunction highlight the need to elucidate the mechanisms of transcriptional regulation of mismatch repair genes. In this study, we examined conserved promoter sequences upstream of MSH2 to decipher the regulatory elements directing the gene’s cell-cycle periodicity and DNA damage-induced expression. Our results show that Msh2 regulation during the cell cycle and in response to DNA damage regulation occurs at the transcriptional and post-transcriptional levels.
2. Materials and methods
2.1. Microbial and molecular manipulations
Strains (Table 1) and plasmids (Table 2) used in this study are detailed below. Microbial manipulations were conducted according to established procedures [27,28]. Polymerase chain reactions (PCR) using primers listed in Supplementary Table 1 were performed as detailed elsewhere [27], and unless otherwise noted, 25 PCR cycles were performed as follows: 94 °C for 15 s, 54 °C for 15 s, and 72 °C for 1 min, followed by 10 min at 72 °C. Yeast colony PCR was performed as described previously [29]. Plasmid DNA was isolated from E. coli according to the QIAprep Spin Miniprep protocol (Qiagen Inc., Valencia, CA). Plasmid and genomic DNA was isolated from S. cerevisiae using published procedures [30]. Diagnostic restriction endonuclease digests of plasmid DNA were performed according to the manufacturer’s specifications (New England Bio-labs, Inc., Beverly, MA), and digested samples were examined by analytical agarose gel electrophoresis [27]. As required, DNA was excised from agarose gels and extracted using the GENECLEAN Spin Extraction Kit (Bio101, Carlsbad, CA).
Table 1.
Strains used in the study.
| Strain | Relevant Markers | Source |
|---|---|---|
| AGY70 | MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 msh2Δ::LEU2 RAD5 | [36] |
| AGY1052 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mbp1Δ::KanMX4 | [34] |
| AGY1053 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 swi4Δ::KanMX4 | [34] |
| AGY1054 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 swi6Δ::KanMX4 | [34] |
| AGY798 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | [34] |
| AGY799 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 msh2Δ::KanMX4 | [34] |
| AGY75 | MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 msh2Δ::LEU2 RAD5 [pSH44,TRP1] | [36] |
| AGY220 | MATα ade2-1 trp1-1 ura3-1 his3-11,15 RAD5 | This study |
| AGY665 | MATα ade2-1 trp1-1 ura3-1 his3-11,15 RAD5 MSH2-MYC::KanMX6 | This study |
| AGY1055 | MATα ade2-1 trp1-1 ura3-1 his3-11,15 RAD5 PROM2 MSH2-MYC::KanMX6 | This study |
| AGY1056 | MATα ade2-1 trp1-1 ura3-1 his3-11,15 RAD5 PRSF1 MSH2-MYC::KanMX6 | This study |
| AGY1057 | MATa pol3-01 msh2Δ::LEU2 leu2-3,112 his3Δ200 trp1-1 ura3-1 [MSH2-MYC:: KanMX6 URA3 CEN/ARS] | This study |
All strains are derived from W303 except for the yeast knock-out collection strains (AGY798-9, AGY1052-4) and the pol3-01 strain (see Section 2). The W303 strains were confirmed to be wild-type at the RAD5 locus by PCR and at the CAN1 locus by canavanine resistance assays.
Table 2.
Plasmids used in this study.
| Plasmid | Strain | Relevant yeast markers | Source |
|---|---|---|---|
| pRS415 | CEN/ARSLEU2 | [32] | |
| pRS413 | CEN/ARSHIS3 | [32] | |
| pSH44 | PLEU2 -(GT)16.5 -URA3TRP1/ARSCEN11 | [37] | |
| pMSH2-218 | AG530 | PMSH2(218) -MSH2CEN/ARSHIS3 | This study |
| pPMSH2-luc | AG531 | PMSH2 -luc CEN/ARSLEU2 | This study |
| pPMSH2ccb−-luc | AG532 | PMSH2ccb−-luc CEN/ARSLEU2 | This study |
| pPADH1-luc | AG533 | PADH1 -lucCEN/ARSLEU2 | This study |
| pMSH2 | AG17 | PMSH2 -MSH2-HA CEN/ARSHIS3 | [36] |
| pMSH2ccb- | AG534 | PMSH2ccb−-MSH2::HA CEN/ARSHIS3 | This study |
| pMSH2upAUG- | AG535 | PMSH2upAUG−-MSH2CEN/ARSLEU2 | This study |
| pMSH2upUAA- | AG536 | PMSH2upUAA−-MSH2CEN/ARSLEU2 | This study |
| pPMSH2(165) luc | AG570 | PMSH2(165) -lucCEN/ARSLEU2 | This study |
| pPMSH2(113) luc | AG571 | PMSH2(113) -lucCEN/ARSLEU2 | This study |
| pPMSH2(69) luc | AG572 | PMSH2(69) -lucCEN/ARSLEU2 | This study |
| pPROM2-MSH2 | AG537 | PROM2 -MSH2::HA CEN/ARSLEU2 | This study |
| pPRSF1-MSH2 | AG538 | PMSH2 -MSH2::HA CEN/ARSLEU2 | This study |
| pPROM2-MSH2-YIp | AG539 | PRSF1 -MSH2 URA3 | This study |
| pPRSF1-MSH2-YIp | AG540 | PRSF1 -MSH2 URA3 | This study |
| pMSH2-MYC | AG541 | MSH2-MYC::KanMX6CEN/ARSURA3 | This study |
| pGAL-MSH2 | AG122 | PGAL10 -MSH2-HA 2μ HIS3 | [36] |
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.dnarep.2012.11.002.
2.2. Plasmid and strain construction
Most of the plasmids (Table 2) created for this analysis were engineered by in vivo homologous recombination in yeast [31]. Briefly, lithium acetate-mediated transformation [29] was used to introduce DNA fragments with homologous ends into S. cerevisiae cells. Plasmid DNA from transformants shown to harbor the desired constructs (as demonstrated by diagnostic PCR) were introduced into E. coli, extracted, and confirmed by restriction endonuclease digestion and sequencing (GENEWIZ Inc., South Plainfield, NJ). The specific details of each construct are given below, and primer sequences are found in Supplementary Table 1.
2.2.1. pPMSH2-luc
The luciferase coding sequence (luc CS) was amplified from pPROLar.A22-luc (Clontech, Palo Alto, CA) using PCR primers MSH2luc 5′ and vecluc 3′. The 218-bp intergenic region upstream of MSH2 (PMSH2) was amplified with primers vecMSH2 5′ and MSH2luc 3′, using pMSH2 as the template. AGY798 cells were transformed with luc CS, PMSH2, and BamHI-linearized pRS415 [32] to produce pPMSH2-luc.
2.2.2. pPADH1-luc
As a positive control for luciferase activity, a plasmid was constructed containing the constitutively active ADH1 promoter (PADH1) driving luciferase expression. PADH1 was amplified from genomic DNA using primers vecADH 5′ and ADHluc 3′, and AGY798 cells were transformed with PADH1, luc CS, and BamHI-linearized pRS415 to create pPADH1-luc.
2.2.3. pMSH2ccb- and pPMSH2ccb−-luc
A mutagenic oligonucleotide (ΔMCB, Supplementary Table 1) was used to disrupt conserved cell-cycle elements of the MSH2 promoter upstream of the wild-type gene (pMSH2) or the luciferase gene (pPMSH2-luc) using site-directed mutagenesis [33]. The primer was designed to scramble the elements while maintaining the nucleotide composition of the conserved region. Mutagenesis was confirmed by DNA sequencing.
2.2.4. pMSH2-AflII
An AflII site immediately upstream of the MSH2 coding sequence in pMSH2 was introduced by site-directed mutagenesis using the primer MSH2AflII. This site allowed for excision of the plasmid’s endogenous MSH2 promoter and replacement with a smaller promoter. Introduction of the restriction site (confirmed by AflII digestion and sequencing) did not produce a detectable mutator phenotype, as pMSH2-AflII fully complemented the mismatch repair defect of an msh2Δ strain (data not shown).
2.2.5. pMSH2-218
pMSH2-AflII was digested with AflII and XhoI, and the ~8-kb vector backbone was excised and purified from an agarose gel. The 218-bp MSH2 promoter (PMSH2) was amplified from pMSH2 using PCR primers vecMSH2Afl 5′ and MSH2PMSH2 3′. The resulting plasmid, pMSH2-218 fully complemented the mismatch repair defect of an msh2Δ strain (see Fig. 1).
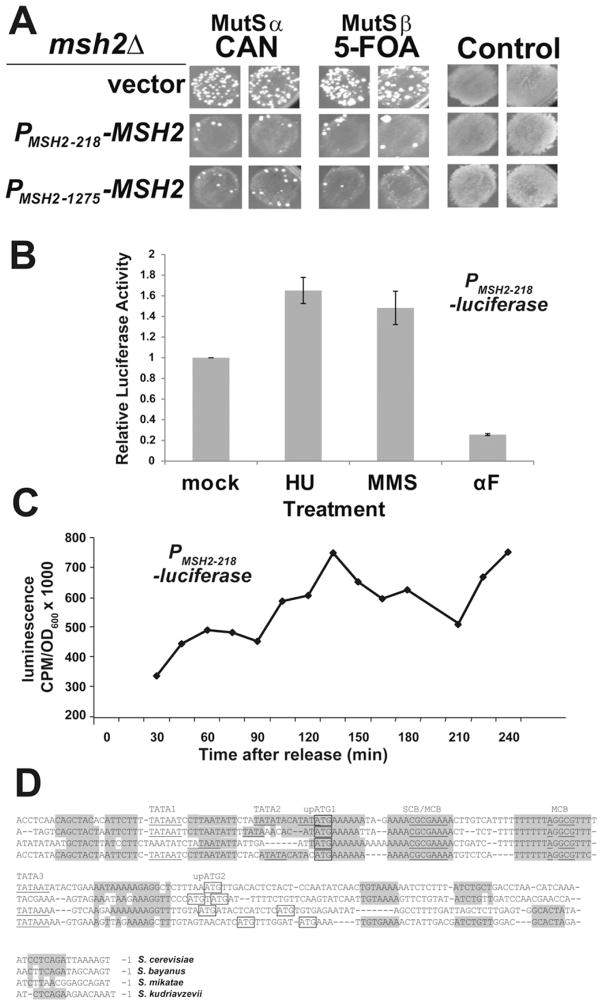
Fig. 1.
Defining the minimal MSH2 promoter. (A) The 218-bp intergenic promoter region upstream of MSH2 is sufficient to confer mismatch repair proficiency. Single msh2Δ transformants harboring the microsatellite instability reporter construct pSH44 (AGY75) and either pRS413 (vector), pMSH2-218 (PMSH2-218 -MSH2), or pMSH2 (PMSH2-1275 -MSH2) were patched onto selective plates and replica printed onto canavanine (CAN), 5-fluororotic acid (5-FOA) and control plates to assess cell viability and plating efficiency. Elevated rates of resistance to 5-FOA reflects the failure to repair polymerase slippage at the dinucleotide tract fused upstream of the URA3 gene on a resident reporter construct [37] associated with Msh2/Msh3 or MutSβ function; whereas, an increased rate of resistance to canavanine represents a failure to repair singe base pair mismatches or single nucleotide insertions/deletions in the CAN1 coding sequence [81] associated with Msh2/Msh6 or MutSα function. (B) The DNA damage responsive element is encoded within the 218-bp intergenic region upstream of MSH2. Exponential phase wild-type yeast cells (AGY798) harboring a plasmid with the 218-bp minimal MSH2 promoter driving luciferase expression (PMSH2-218-luciferase) were mock treated or treated with 0.1 M HU, 0.04% MMS, or 10 μg/ml αF for 1.5 h prior to luciferase analysis. Results were reproduced in 8 independent experiments for HU, MMS, and 3 independent experiments for the αF treatment. Relative luciferase activity was calculated by dividing by light emission of mock-treated cells. Error bars represent standard error of the mean. (C) The minimal MSH2 promoter contains cell-cycle periodicity function. Yeast cells described above were sequentially arrested with nocodazole and αF and released into fresh medium. Every 15 min beginning at t = 30 min, aliquots were sampled for luciferase activity. CPM represents counts per million. The yeast doubling time is ~90 min (not shown). (D) Interspecies alignment highlighting conserved MSH2 promoter elements. The promoter sequences of S. cerevisiae MSH2 and homologs were compared using the fungal alignment viewer [50]. Conserved elements are shaded. Putative TATA boxes (1–3), SCB, and MCB elements are indicated and nucleotides matching the consensus sites are underlined. The ATG codons for the uORFs (upATG1,2) are boxed.
2.2.6. pMSH2upAUG- and pMSH2upUAA
The primers MSH2-UP-ATG and MSH2-UP-TAA were designed to disrupt the start and stop codons, respectively of the upstream open reading frame, using site-directed mutagenesis [33]. Muta-genesis was confirmed by DNA sequencing.
2.2.7. pPMSH2(165)-luc, pPMSH2(113)-luc, and pPMSH2(69)-luc
5′ deletion analysis of the MSH2 promoter was performed by PCR with upstream primers DEL3 5′, DEL4 5′, and DEL5 5′ and downstream primer MSH2luc 3′ (Supplementary Table 1) using homologous recombination, as described above for pPMSH2-luc.
2.2.8. swi6Δ, swi4Δ, and mbp1Δ
kanMX4 junctions in the yeast deletion collection knockout strains [34] were confirmed by polymerase chain reaction using primers ~300-bp upstream and downstream of each deleted gene (swi6 5′ and swi6 3′; swi4 5′ and swi4 3′; mbp1 5′ and mbp1 3′) and primers complementary to the kanMX4 cassette (kan 5′ and kan 3′).
2.2.9. AGY665 (MSH2- MYC::kanMX6)
The C terminal-coding region of MSH2 in strain AGY220 was fused to a kanamycin marked MYC epitope tag (MYC::kanMX6) using a PCR-based method as described previously [35]. A 20-cycle PCR reaction of 1 min at 94 °C, 1 min at 50 °C, and 2.5 min at 68 °C, followed by a final 10-min extension at 68 °C, was used to amplify the MYC::kanMX6 cassette from pFA6a-13Myc-kanMX6 [35], using MSH2TAG5 and MSH2TAG3 primers. The strain was confirmed by PCR of the fusion junctions and the functionality of the fusion protein was tested in a canavanine drug sensitivity assay.
2.2.10. AGY1055 (PROM2-MSH2-MYC::kanMX6) and AGY1056 (PRSF1-MSH2-MYC::kanMX6)
Promoter swamp integrations were accomplished by first generating centromere-based plasmids in which the 218-bp region between SPO21 and MSH2 was replaced with the 562 intergenic region upstream of RSF1 (pPRSF1-MSH2) or the 460 bp region upstream of ROM2 (pPROM2-MSH2). The constructs were made by combining HindIII-linearized pRS415 [32] and PCR fragments generated using primers vecSPO and MSH2up and primers MSH2dwn and vecMSH2 along with either primers PRSF1up and PRSF1dwn for the PRSF1 region or the PROM2up and PROM2dwn for the PROM2 region. After verification of the constructs, fragments containing the promoter swaps were subcloned into a URA3-based yeast integrative plasmid (pRS406) using NotI and XhoI to generate pPRSF1-MSH2-YIp and pPROM2-MSH2-YIp. These YIp plasmids were linearized with HindIII to integrate into AGY665 (MSH2-MYC::kanMX6). The two-step integration of the promoter swap mutants AGY1055 (PROM2-MSH2) and AGY1056 (PRSF1-MSH2) was verified by PCR.
2.2.11. pMSH2-MYC
This construct expresses MYC-tagged Msh2 from a URA3 CEN-based plasmid. The plasmid was constructed in vivo using 3 PCR products and a BamHI linearized pRS416 [32]. The first fragment was amplified from wild-type genomic DNA with primers vecMSH2 5′ and 5341(−) to produce a 2.1 kb fragment containing the 218-bp MSH2 promoter and 1.9 kb of the N-terminal MSH2 coding sequence. The second fragment was amplified from wild-type genomic DNA using primers 5200(+) and 6260(−) to produce a 1-kb fragment containing MSH2 coding sequences with flanking sequence homology to fragments 1 and 3. The third fragment was amplified from AGY665 genomic DNA with primers MSH2-12(+) and vecMSH2utr3′ to generate a fragment of ~2.7 kb containing the MYC-tagged MSH2 C-terminal coding region, the kanMX6 marker and 126 bp of the 3′ untranslated region of MSH2. The plasmid complemented the mismatch repair defect of a msh2Δ strain and rescued the pol3-01 msh2Δ synthetic lethality.
2.2.12. AGY1057 (pol3-01 msh2Δ+ pMSH2-MYC)
AGY70 (MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 msh2::LEU2 RAD5) + pMSH2-MYC was mated to EAY575 (MATα ura3-52 leu2Δ1 his3Δ200 pol3-01; generously provided by Eric Alani, Cornell University), to generate AGY1057. Synthetic lethality was confirmed on plates containing 5-fluoro-orotic acid (5-FOA).
2.3. Mismatch repair and synthetic lethality assays
DNA mismatch repair assays were performed to test for function of the promoter mutants. Constructs were used to transform AGY75, a msh2Δ reporter strain [36], and transformants were tested for DNA mismatch repair using quantitative and qualitative assays as described previously [37–39].
Plasmid shuffle assays to test the mismatch repair in a pol3-01 background employed AGY1057, a msh2Δ pol3-01 + pMSH2-MYC strain transformed with additional plasmids including pMSH2, pP MSH2ccb−-MSH2, or pRS413. Two-fold serial dilutions of exponentially growing cells in synthetic medium lacking histidine were plated on plates containing synthetic medium lacking histidine, lacking histidine supplemented with 5-FOA, or lacking histidine supplemented with 5-FOA and 0.04% MMS. Plates were grown at 30 °C for 2 days.
2.4. Luciferase assays
Cells harboring MSH2 promoter-luciferase fusion constructs or empty vector (pRS415) were grown to mid-exponential phase (OD600 ~ 0.4) in synthetic medium lacking leucine (−LEU), and luciferase activity was assayed in a protocol modified from Vieites and colleagues [40]. Briefly, cells were harvested, washed twice with dH2O, and resuspended in 10 mM potassium phosphate buffer (pH 7.0). Optical density at 600 nm (OD600) was determined by spectrophotometry, and ~3 OD600 of cells were added to prewarmed (30 °C), oxygenated (15-s vortex) sodium citrate buffer (pH 5.5). D-Luciferin (20 mM stock) was added to a final concentration of 2.0 mM. Cells were briefly vortexed, moderately shaken for 15 min, and vortexed again, and luciferase activity was measured in a Wallac 1409 liquid scintillation counter (wave-length of emission = 562 nm). OD600 of the final reaction chamber was determined to allow cell density adjustments, as described below.
In assays with drug-treated cells, mid-exponential cultures were treated with 10 μg/ml alpha factor (αF, Princeton Syn/Seq Facility, Princeton University), 0.1 M hydroxyurea (HU, Sigma–Aldrich, Co., St. Louis, MO), or 0.04% v/v methyl methane sulfonate (MMS, Sigma–Aldrich, Co.) for 1.5 h, followed by luciferase assays as described above. Mock-treated cells and cells harboring pRS415 were similarly prepared. Cell-cycle arrest (>80% and >60% for αF and HU, respectively) was confirmed by phase contrast microscopy. Serial dilutions of MMS-treated cells were plated on −LEU to confirm the efficacy of the MMS stock and, at lower concentrations, to confirm the absence of significant cell killing.
Data from luciferase assays were analyzed as follows. Values of luciferase activity generated by a Wallac 1409 were divided by the final OD600 of the reaction chamber to yield a normalized value of luciferase activity (observed light emission/OD600). The light emission of WT cells harboring pRS415 was used as a background control for luminescence. Control experiments demonstrated that light emission from cells harboring pRS415 was essentially identical in all strain backgrounds and that luminescence in pRS415-transformed cells did not vary significantly with drug treatment (data not shown). Luciferase activity in cells harboring a plasmid with a transcription terminator cloned upstream of the luciferase start codon was not significantly higher than that of cells harboring empty pRS415, confirming the absence of significant background luciferase expression. Overall, luciferase activity was expressed as: (observed light emission/OD600) – background light emission. Due to large inter-trial variation in the absolute value of light emission measurements, values in each trial were normalized to emission of wild-type and/or mock-treated cells, as appropriate.
2.5. Cell-cycle analysis of MSH2 promoter activity
To determine the cell-cycle periodicity of the wild-type promoter, exponentially growing AGY798 cells harboring pPMSH2-luc and pPADH1-luc were arrested with nocodazole (1.5 μg/ml final concentration, 4 h), released into αF (10 μM final concentration, 1.5 h), and released from αF arrest into fresh −LEU medium. Phase contrast microscopy confirmed ~50% arrest (large-budded phenotype) with nocodazole treatment and >80% shmooing with αF treatment. Time points were taken at 0 min and every 15 min for 5.5 h starting at t = 30 min. At each time point, 4 ml of cells from each of the three cultures were harvested and assayed for luciferase activity as described above. The OD600 of each final reaction chamber was used to adjust light emission values (described above). After OD600 determination, cells were fixed in 70% ethanol, and nuclei were visualized by staining with 4′,6′-diamidino-2-phenylindole (DAPI) [41]. Cell cycle synchrony was confirmed by fluorescence microscopy. Since ADH1 expression is known to be independent of the cell cycle [16], light emission from cells harboring pPADH1-luc was normalized to the value at t = 0 min by calculating a correction constant for each time point. Emission from cells harboring pPMSH2-luc was adjusted using the correction constant calculated for the given time point.
For the cell synchrony experiments with the wild-type and cell-cycle box scrambled MSH2 promoters driving luciferase, a bar1 protease mutant was used to increase the sensitivity αF. Cells were arrested and released at 18 °C to slow the progression of the cell cycle. After release from αF-induced G1 arrest, aliquots of cells were analyzed with a Promega Glomax 96-well Microplate Luminometer to assess light emissions. Cell synchrony was verified by flow cytometry.
2.6. Primer extension
RNA was isolated from yeast cells according to established procedures (TRIzol Reagent RNA Isolation, Invitrogen Life Sciences, Carlsbad, CA). Isolated RNA was run on a 1% agarose gel and visualized with ethidium bromide to confirm RNA integrity, as indicated by the presence of distinct ribosomal RNA bands. The oligonucleotide primer (MSH2-5′REV) was labeled with γ32P-ATP (activity >7000 Ci/mmol; ICN Biomedicals, Inc, Irvin, CA) following published protocol [27]. Purification of the labeled oligonucleotide was accomplished using a ProbeQuant G-50 micro column (GE Healthcare Life Sciences, Piscataway, NJ). Primer extension using the labeled oligonucleotide was performed on RNA isolated from AGY798 (wild-type) and AGY799 (msh2Δ) using the First Strand cDNA Synthesis Using SuperScript II RT protocol (Invitrogen Corporation, Carlsbad, CA). DNA was then precipitated using 0.6 volumes 5 M ammonium acetate and 3 volumes 100% ethanol. The pellet was washed with 70% ethanol, air dried, resuspended in DNA sample buffer, and heated at 65 °C for 5 min. DNA was fractionated on a polyacrylamide sequencing gel following published protocol [27] and using Sequagel reagents according to the manufacturer’s specifications (final concentration: 8.3 M urea/6% (w/v) acrylamide/1× Tris–borate–EDTA; National Diagnostic, Atlanta, GA).
Sequencing reactions of MSH2 were performed using the same oligonucleotide used in the primer extension. The sequencing reaction products were run simultaneously on the gel along with the primer extension products in order determine their location and length along the MSH2 sequence. The sequencing was performed with the labeled oligonucleotide using the Amersham LIFE SCIENCE Sequenase Version 2.0 sequencing kit (T7 Sequanase Chain-Termination Sequencing Protocol; Amersham Life Sciences, Arlington Hts, IL).
2.7. Immunoblotting analysis
Yeast protein extracts from exponential cells grown for 1.5 h in the presence or absence of 0.04% MMS were prepared as described elsewhere [29,42], using ~2 OD600 of each culture. Extracts were fractionated by discontinuous sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) [27] using the BioRad Mini-Protean II Cell, a 4% acrylamide stacking gel (pH 6.8), and a 7% acrylamide separating gel (pH 8.8). Overnight transfer to nitrocellulose was confirmed by staining with Ponceau Red (Sigma–Aldrich, St. Louis, MO) [27], and the membrane was blocked for 1–24 h with 5% nonfat dry milk (NFDM) in Tris-buffered saline +0.1% Tween 20 (TBS-T). Immunodetection of hemagglutinin (HA)-tagged Msh2 was performed by Enhanced Chemiluminescence (ECL) western blotting (Amersham Biosciences, Piscataway, NJ) using a 1:2500 dilution of mouse α-HA monoclonal primary antibody (12CA5; Princeton Monoclonal Facility) and a 1:2500 dilution of donkey α-mouse IgG horse radish peroxidase (HRP)-conjugated secondary antibody (Amersham Biosciences; 45 min). After exposure on X-OMAT XAR-5 Kodak film (1–5 min), the membrane was washed twice with TBS-T (10 min) and reprobed for Kar2 using rabbit α-Kar2 polyclonal primary antibody (M. D. Rose Laboratory, Princeton University; 1:10,000 dilution) and goat α-rabbit IgG-HRP secondary antibody (GE Healthcare Life Sciences). Quantification of band intensity was performed using the public domain Image J program. The fold-induction over wild-type Msh2 levels for the strains harboring plasmids with the upstream open reading frame start and stop codons were based on 3 independent experiments.
2.8. Msh2 turnover assays
The methods for construction of MSH2 under the GAL10 promoter (PGAL10) were described previously [36]. Cells expressing the PGAL10 fusion were grown to logarithmic phase in synthetic medium with 2% galactose. The culture was split and glucose (2%) was added to each culture to repress synthesis of MSH2. One culture was also exposed to MMS (0.04%) while the other was mock-treated. At time intervals of 0 h, 2 h, and 4.0 h after repression of synthesis, ~3 × 107 cells were processed for immunoblotting. Protein extracts were subjected to chemiluminescence immunoblotting methods to detect Msh2 variant and a loading control.
During the shut-off experiment cell division occurred complicating the turnover analysis. Because transcription of MSH2 was repressed, each cell division diluted the Msh2 by half. Therefore, the decrease in Msh2 signal is a combination of degradation and cell division. Additionally, cells treated with MMS grew at a slower rate than those cultures mock treated. Thus, correction for cell growth was required for a more accurate comparison of the turnover rate. To correct for the dilution due to cell division, we analyzed the turnover bands by densitometry, set the 0 h point as 100% starting protein, and subsequent time points were divided by this initial value to produce a percentage of Msh2. A trendline was imposed on this data to identify the exponential rate of protein decay (due to both dilution and cellular degradation, e−Rt). We then plotted the OD600 of the cultures versus time and identified the exponential rate at which the cells were growing (eKt) during the experiment. To isolate the turnover due purely to cellular degradation, these two equations were multiplied to find the corrected values (e(K−R)t).
3. Results and discussion
3.1. The 218-bp intergenic region upstream of MSH2 contains cell-cycle and DNA damage responsive promoter elements
To define the regulatory elements controlling MSH2 expression, the minimal region necessary for wild-type expression patterns of MSH2 during mitotic growth was determined using upstream regions of varying lengths. Expression was driven either by the 1275-bp region partially spanning the C-terminal coding sequence of SPO21 (immediately upstream of MSH2 on chromosome XV) or by the 218-bp intergenic region upstream of MSH2 and downstream of SPO21. These constructs served as conservative and minimal estimates of MSH2 promoter length, respectively. To determine whether the 218-bp promoter was sufficient for effective mismatch repair in vivo, the spontaneous mutation rates of msh2Δ cells with MSH2 expression under the control of the two promoters described above were assessed qualitatively using negative selection papillation assays. Cells harboring the vector produced considerable papillation on the negative selection plates, indicative of the expected strong defect in mismatch repair. In contrast, MSH2 was expressed to sufficient levels from both the 218-bp and a larger 1275-bp promoter to complement the mismatch repair defect of an msh2Δ strain (Fig. 1A). Thus, the 218-bp intergenic region upstream of MSH2 was sufficient to confer effective mismatch repair function and was therefore used in subsequent analyses.
To allow rapid quantification of MSH2 promoter activity, we generated plasmids in which the 218 bp region upstream of MSH2 drives expression of firefly luciferase. The promoter fusions were engineered at the start codon such that the luciferase open reading frame replaced the MSH2 coding sequence (PMSH2-luciferase), thereby preserving the 5′ UTR regulatory elements.
To confirm that luciferase activity is an accurate proxy for MSH2 expression and to verify previously assembled microarray data, the luciferase assay described above was used to monitor MSH2 promoter activity in response to treatment with hydroxyurea (HU), methyl methanesulfonate (MMS), and alpha-factor (αF), substances known to influence MSH2 mRNA levels. To assess the effects of these agents on MSH2 promoter activity, cells expressing PMSH2-luciferase were grown to mid-exponential phase and mock-treated or treated with 0.1 M HU, 0.04% MMS, or 10 μg/ml αF for 1.5 h prior to analysis. Luciferase activity revealed a ~1.7-fold increase in promoter activity upon treatment with HU as compared to mock-treated cultures (Fig. 1B). This increase is similar to that observed in microarray studies (~1.4 fold, [43]). Similarly, we observed a ~1.5-fold increase in luciferase activity in response to treatment with 0.04% MMS (Fig. 1B), in accordance with several sets of microarray data [17,44]. αF treatment resulted in decreased expression compared to the peak during S-phase (~26% of the highest value in S-phase), consistent with previous observations in genome wide analyses [45].
The elements controlling cell-cycle periodicity were also encoded within the 218-bp intergenic region found immediately upstream of MSH2 (Fig. 1C). Cells expressing the luciferase gene under the control of the MSH2 promoter were arrested in the G1 phase of the cell cycle, released from arrest, and assayed over time for luciferase activity. The cell-cycle periodicity conferred by the MSH2 promoter is reflected in the peaks and valleys of light emission with the approximately 90-min separations consistent with the doubling time of yeast. Taken together, these results indicate that the 218-bp intergenic region upstream of MSH2 contains HU-, MMS-, and cell-cycle-responsive elements of the MSH2 promoter.
Comparative genomics of related species is a powerful technique for identifying conserved regulatory elements [reviewed in 46]. Interspecies genome alignments were used to search for conserved sequences potentially involved in the transcriptional regulation of MSH2 (Fig. 1D). Specifically, the Fungal Sequence Alignment [47–49] through the Saccharomyces Genome Database [50] was utilized. The analysis revealed conserved regions, several potential TATA boxes, two upstream small open reading frames (uORFs), and two known cell-cycle regulatory elements. The two cell-cycle elements are a MluI cell cycle box (MCB) and an overlapping Swi6/4 cell cycle box (SCB)/MCB box. MCBs and SCBs are common elements upstream of many genes that exhibit peak expression at the G1/S boundary [reviewed in 51]. Together, these findings define a 218-bp minimal promoter region containing several elements that might contribute to the cell-cycle and DNA damage regulated expression of MSH2.
3.2. A small upstream open reading frame present in the MSH2 transcript modulates MSH2 expression during mitotic growth
To determine whether the conserved regions of the MSH2 promoter were present in the MSH2 transcript, we used primer extension analysis to map the 5′ end of the MSH2 transcript (Fig. 2A). The mapping was performed on RNA isolated from wild-type and msh2Δ strains. Two transcript start sites were identified. The longer transcript initiates in a poly-T run between the SCB and MCB boxes (Fig. 1D). The shorter transcript initiates downstream of both cell cycle boxes and a putative TATA box. The primer extension analysis for the transcript initiating closer to the MSH2 ATG is in agreement with genome-wide tiling array experiments mapping the yeast transcriptome [52] (Fig. 2B and C). Interestingly, the 5′ end mapping confirmed that a conserved and potentially regulatory open reading frame is present within MSH2 transcripts (Fig. 2D).
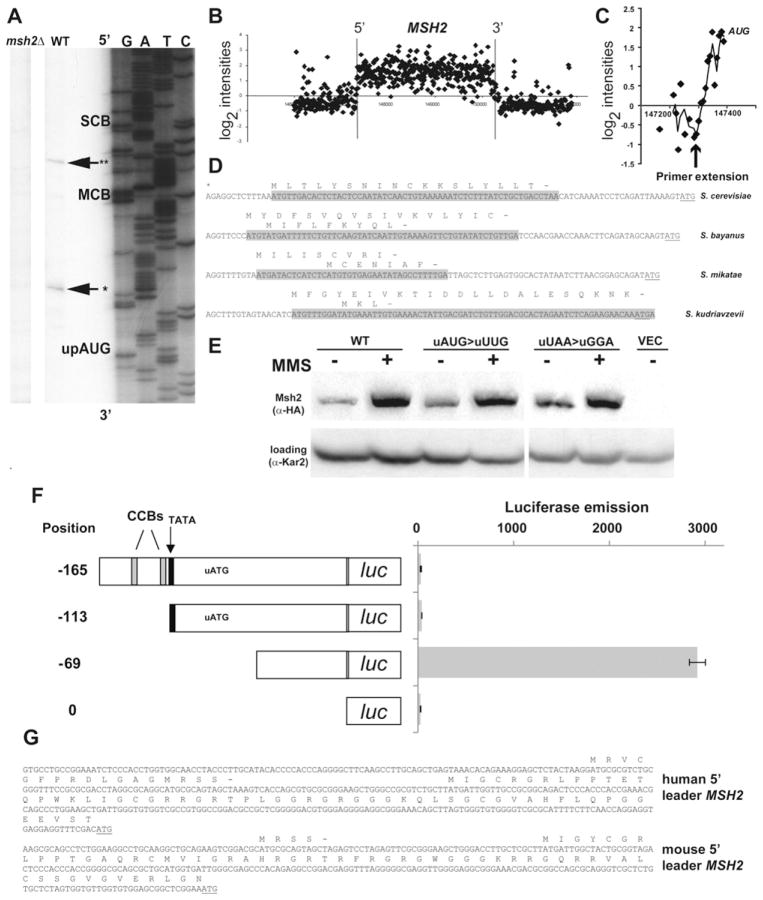
Fig. 2.
An upstream open reading frame is encoded within the mRNA of MSH2. (A) Primer extension analysis of MSH2 mRNA. The 5′ end of the MSH2 transcript was mapped using RNA from a wild-type (WT, AGY798) and an msh2Δ (AGY799) strain. Parallel sequencing reactions were performed with the same labeled primer used in the primer extension. Potential regulatory regions are indicated to the right and correspond to the highlighted regions in Fig. 1. The reverse complement of the sequence in the previous figure is shown with GATC indicated above the lanes. The position of the 5′ end of the transcripts (*, **) are emphasized with an arrow. The start of the upstream open reading frame (uORF) is indicated. (B) A high resolution map of the MSH2 transcript. David et al. [80] mapped the entire yeast transcriptome including the MSH2 locus. Data for MSH2 was downloaded from the ArrayExpress database, accession no. E-TABM-14. The plot displays the log2 normalized expression level (y-axis) along genomic coordinates (x-axis in bp). Each dot corresponds to a probe on the tiling array and the vertical lines represent the boundaries of the MSH2 open reading frame. The background threshold (y = 0) is shown as a horizontal line. (C) The tiling array data and the primer extension analysis are in accordance for the start site of the MSH2 mRNA. The 5′ end of the MSH2 region is magnified to show the individual probe hybridizations from panel B. The start site from the primer extension analysis is indicated with an arrow and the start codon for MSH2 is indicated. (D) A conserved upstream open reading frame is encoded in the mRNA of MSH2. A sequence alignment of the 5′ region upstream of MSH2 of related species is shown. The mapped start site of the transcript (*) closest to the MSH2 ATG (underlined) and the conserved upstream open reading frames are indicated above the putative coding region shaded in gray. The MSH2 start codon is underlined. (E) Disruption of the upstream ORF increases Msh2 protein levels during mitotic growth. Strains with msh2 deleted (msh2Δ, AGY75) were transformed with a vector (VEC), the pMSH2 plasmid expressing MSH2 (WT), or mutagenized pMSH2 where the upstream open reading frame was disrupted by changing the start codon (uAUG>uUUG) or the stop codon (uUAA>uGGA). Exponentially growing cultures were grown for 1.5 h in the absence (−) and presence (+) of MMS. Protein extracts were analyzed by immunoblotting with α-hemaglutinin (HA) to detect Msh2 or with α-Kar2p as a loading control. (F) Deletion of the upstream ORF in the 5′ leader sequence increases luciferase expression. Strains expressing various 5′ upstream regions of MSH2 fused to the luciferase coding sequence (luc). The start position of the leader is indicated with respect to the start codon. Salient features (TATA, black box; cell-cycle boxes, CCBs; and the upstream open reading frame start codon, uATG) are indicated above the schematic diagrams of the promoter. Luciferase emission is counts per minute normalized to optical density. Error bars reflect standard error of the mean. (G) Conservation of upstream open reading frames in the 5′ region of human and mouse MSH2 transcripts. The human and mouse MSH2 uORFs are shown above the sequence. The sequences were obtained from AceView: a comprehensive cDNA-supported gene and transcripts annotation [59].
Fewer than 10% of eukaryotic genes are predicted to contain small open reading frames in their transcript leader sequences. However, a subset of genes involved in cell growth and differentiation contain uORFs [reviewed in 53,54]. The conservation of the uORF in the transcript leader sequence and the verification that the uORF is encoded in the transcript increases the likelihood the uORF plays a regulatory role in translation initiation [reviewed in 55]. To test for a potential inhibitory role of the uORF in the context of the wild-type MSH2 gene, we engineered mutation in the ATG codon (ATG>TTG) of the uORF immediately upstream of MSH2. We also mutated the stop codon (TAA>GGA) for the uORF, which results in a continuous reading frame with the MSH2 coding sequence. We observed increases in Msh2 protein levels in mitotic cells for the ATG-mutated construct (1.4 ± 0.3 fold increase, n = 3) and for the stop codon-mutated construct (1.3 ± 0.2 fold increase, n = 2) (Fig. 2E). The MMS-induced levels of Msh2 for all three were indistinguishable, suggesting that the increase in translational efficiency was during mitotic growth and not in response to DNA damage.
To further test whether the upstream ORF might play a role in translational efficiency we employed the luciferase assays to eliminate obscuring post-translational controls associated with the wild-type Msh2 protein (our unpublished observation). We constructed plasmids with various deletions of the 5′ leader sequence of MSH2 upstream of the luciferase coding sequence. The results showed when the upstream ORF start codon was deleted, a significant increase in luciferase expression was observed (Fig. 2F). These findings are consistent with the upstream ORF playing an inhibitory role in translation of downstream open reading frames.
The Cln3 cyclin [56] and the Rok1 RNA helicase [57] are two other yeast cell-cycle-regulated genes that display translational control mediated by uORFs. The presence of conserved regulatory uORFs in organisms ranging from fungi [54] to mammals [58] suggests that this form of translational control is widespread. Interestingly, both the mouse and human MSH2 5′ UTRs contain uORFs [59] (Fig. 2G), consistent with a conservation of regulation.
3.3. SCB and MCB cell-cycle boxes and cell-cycle transcription factors are important for basal MSH2 promoter activity
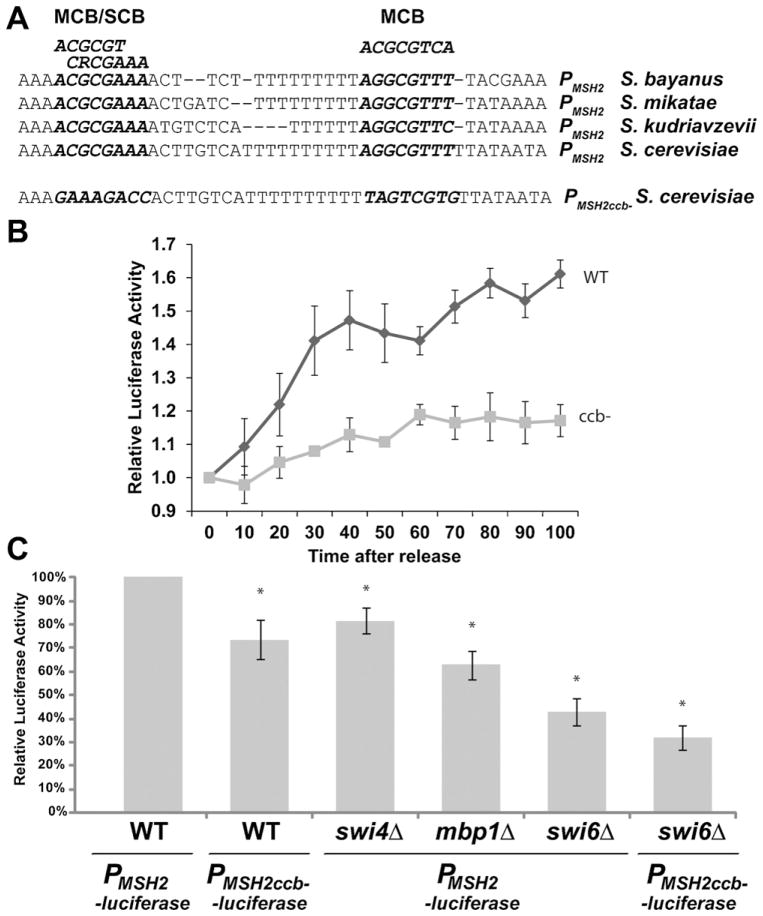
Next, we assessed the role of the DNA-encoded cell-cycle elements in MSH2 expression. Specifically, we used site-directed mutagenesis to scramble the two cell cycle boxes (ccb-) within the MSH2 promoter driving expression of luciferase (PMSH2ccb−-luciferase) (Fig. 3A). Mutagenizing the cell cycle boxes abolished the cell-cycle periodicity associated with the MSH2 minimal promoter region (Fig. 3B). Cells were synchronized and the light emissions from the wild-type and cell cycle box mutant promoter fusions were assessed. The cell synchrony for both strains was confirmed by flow cytometry. In addition to abolishing the cell cycle periodicity, the mutagenesis of these two promoter elements reduced luciferase activity to a level ~65% that of cells harboring a plasmid with a wild-type MSH2 promoter (Fig. 3C, p < 0.02).
Fig. 3.
Effect of deletion of SWI6, SWI4, MBP1, and cell cycle boxes on MSH2 promoter activity. (A) Sequence of the MSH2 promoter region. The conserved MCB/SCB, and MCB sites from related yeast species are indicated. The consensus for the MCB (ACGCGTCA) and SCB (CRCGAAA) are above the alignment. The sequence for the cell-cycle box disrupted promoter mutant (PMSH2ccb−) is also shown, where the two elements (bold and italics) have been scrambled by site-directed mutagenesis. (B) The MSH2 promoter with scrambled cell-cycle boxes no longer displays cell-cycle periodicity. Yeast cells lacking the Bar1 protease harboring PMSH2 -luciferase (WT) PMSH2ccb− -luciferase (ccb-) were arrested with αF and released into fresh medium at 18 °C to slow the cell cycle. Every 10 min beginning at t = 0 min, aliquots were sampled for luciferase activity and for flow cytometry. The luciferase emissions were normalized to the t = 0 time point. Two independent experiments with 3 technical replicates are represented in the figure. Error bars are standard error of the mean. (C) Cell cycle transcription factor deletions reduce MSH2 promoter activity. Normalized light emission of AGY798 wild-type cells (WT), AGY1054 swi6Δ, AGY1053 swi4Δ, and AGY1052 mbp1Δ cells harboring PMSH2 -luciferase as well as WT and swi6Δ cells harboring PMSH2ccb−-luciferase are shown. Results for each strain are the average of three to six independent experiments and are presented as the relative luciferase activity as a percentage of wild-type (WT). Error bars reflect the standard error of the mean. Asterisks indicate p values less than 0.02 when comparing the values to wild-type cells harboring PMSH2 -luciferase.
MCBs are bound by the MluI cell-cycle box binding factor (MBF), a complex composed of Swi6, a transcriptional activator, and Mbp1, a sequence-specific DNA binding protein [60,61]. Additionally, Swi4, as part of the Swi4/Swi6 cell-cycle box binding factor (SBF), binds and activates G1/S genes with SCB-containing promoters [61,62]. To determine whether these transcription factors are important for regulating basal MSH2 expression, we introduced the PMSH2-luciferase reporter construct into swi4Δ, mbp1Δ, swi6Δ and wild-type cells. We found that deletion of SWI4 resulted in a ~20% reduction in luciferase activity (Fig. 3C, p < 0.008). However, there is documented functional redundancy between Swi4 and Mbp1 [61], and in the MSH2 promoter the SCB element overlaps with an MCB element (Fig. 3A). Deletion of MBP1 resulted in a more significant reduction in luciferase activity (to ~60% of WT levels, p = 0.0005, Fig. 3C). The absence of Swi6, important for SBF and MBF function, reduced luciferase activity to ~45% that of wild-type cells (WT vs. swi6Δ p < 0.0001, Fig. 3C). This reduction of promoter activity is not due to a general defect in transcription, as light emission in swi6Δ cells harboring an ADH1 promoter-driven luciferase gene (on plasmid pPADH1-luc) was comparable to expression in wild-type cells harboring the same plasmid (data not shown). The introduction of cell-cycle box deleted promoters into swi6Δ cells did not further reduce light emission to a significant degree when compared to the wild-type promoter (Fig. 3C, p = 0.17), consistent with a model in which Swi6 functions with the partially redundant Swi4 or Mbp1 to control expression through the SCB/MCB sites in the promoter region of MSH2. Swi6 appears to have a stronger effect on the MSH2 promoter and thus may play a more significant role than either Swi4 or Mbp1 because of its role in recruiting chromatin modifying factors to promoter regions [63].
Together, these data demonstrate that the SCB and MCB cell-cycle boxes and cell-cycle transcription factors are important for basal MSH2 promoter activity. The mammalian functional homologues of MBF and SBF are the E2Fs, a family of transcription factors critical in controlling cell-cycle progression and regulating the expression of genes required for DNA replication [reviewed in 64,65]. The hMSH2 promoter contains E2F binding sites at positions −198 and −67, and previous studies have shown that E2F is involved in regulating the cell-cycle-dependent expression of hMSH2 [20]. The identification of these cell-cycle regulatory elements in yeast and mammalian MSH2 promoter regions suggests that conservation between the mismatch repair systems extends to gene expression.
3.4. Disruption of cell-cycle elements upstream of MSH2 results in a defect in mismatch repair
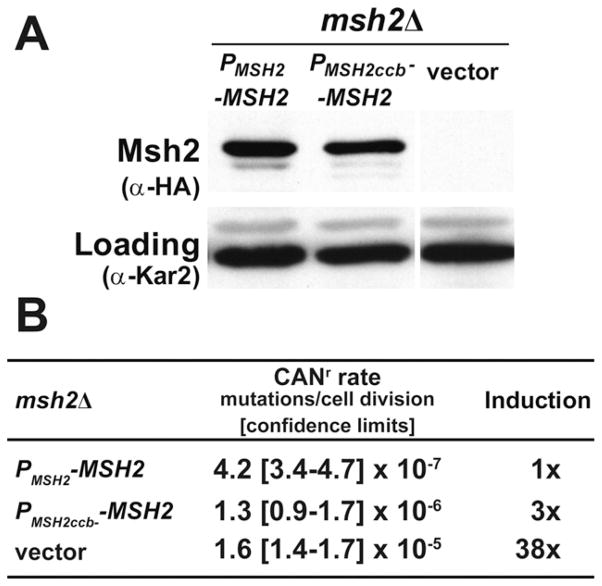
The cell-cycle-regulated expression of MSH2—with a peak at the G1/S boundary—is consistent with the role of Msh2 in post-replicative DNA repair. To determine the effect of cell-cycle regulation on protein levels and mismatch repair function, we assessed steady-state Msh2 protein levels in msh2Δcells harboring a plasmid with the wild-type promoter (PMSH2-MSH2) or the cell-cycle box mutagenized promoter (PMSH2ccb−-MSH2). Results show that the cell-cycle box mutations causes a subtle but reproducible reduction (Fig. 4A) to 56 ± 3% wild-type levels consistent, with the ~40% reduction in MSH2 promoter activity described above (Fig. 3B).
Fig. 4.
Scrambling of cell-cycle elements upstream of MSH2 results in a 3-fold defect in mismatch repair. (A) The expression of Msh2 is slightly reduced when the cell cycle boxes are scrambled. AGY75-derived strains lacking MSH2 (msh2Δ) and harboring either pRS413 (vector), pMSH2 (PMSH2 -MSH2), or a construct where the cell-cycle boxes upstream of MSH2 are scrambled (PMSH2ccb− -MSH2) were grown to exponential phase, and whole-cell protein extracts were analyzed by immunoblot-ting with α-hemaglutinin (HA) to detect Msh2 or with α-Kar2 as a loading control. (B) Disruption of MSH2’s cell-cycle boxes results in a 3-fold increase in mutation rate. The mutation frequencies of the strains described above were assessed using cana-vanine resistance assays (CANr) and expressed in mutations per cell division. 95% confidence limits are shown between the brackets. The fold induction of mutation rate (induction) was calculated by dividing the value by the wild-type rate.
To assess the ability of PMSH2ccb MSH2 to complement the mismatch repair defect of an msh2Δ strain, we used median-based fluctuation tests to obtain a quantitative measure of spontaneous mutation rate. The results revealed a 3-fold increase in the mutation rate at the CAN1 locus as compared to msh2Δ cells complemented by wild-type MSH2 under its endogenous promoter (Fig. 4B). This relatively subtle effect of cell-cycle box mutagenesis on mismatch repair compared to the ~38-fold increase in mutation rate observed with msh2Δ cells harboring and empty vector was nevertheless significant (based on the 95% confidence limit interval, Fig. 4B).
We speculate that the mutator phenotype is a consequence of the loss of cell-cycle regulation rather than lower protein levels based on our previous characterization of msh2 missense variants [36]. These missense alleles of msh2 retained their cell-cycle boxes and showed a variety of phenotypes, including differing protein levels and mismatch repair defects. Among the panel of variants, we have two examples with protein levels ~65% of wild-type that displayed no significant mutator phenotype [36]. Further support for the model comes from the observation we did not detect a significant increase in the mutation rate in the mbp1Δ and swi4Δ (not shown), strains in which cell cycle periodicity is typically retained because of the significant redundancy [66].
3.5. Cell-cycle boxes are not required for the induction of MSH2 in response to the DNA damaging agent methyl methane sulfonate
Previously studies have reported a link between cell-cycle regulation and DNA damage-induced expression of certain G1/S-induced S. cerevisiae genes [19,67–69]. Likewise, cell-cycle transcription factors have been proposed to regulate expression of DNA damage response genes in mammalian cells [20]. Thus, a parsimonious model emerges in which cell-cycle transcription factors are responsible for both cell cycle-regulated and DNA damage-induced expression of DNA synthesis and repair genes, allowing DNA repair outside of S phase [70].
To test whether MSH2 expression fits this model, we analyzed the role of the cell-cycle boxes in response to DNA damage using a strain that is dependent upon DNA mismatch repair for survival. The strain encodes a mutation in DNA polymerase δ (encoded by POL3), causing a defect in the proofreading function. Previous studies have shown that this allele of POL3 (pol3-01) causes cells to die at elevated temperatures in combination with mutations in known DNA mismatch repair genes [71–77]. We chose to use this system because of the high degree of sensitivity capable of detecting even modest (6-fold) mutator phenotypes. Additionally, because we were using a mutagen (MMS), we did not want to obscure the results by using mutation frequency assays.
In our strain background, the combination of the pol3-01 allele and a deletion of MSH2 (msh2Δ pol3-01) is lethal at all temperatures. The pol3-01 msh2Δ strain is viable when a covering plasmid expressing Msh2 is maintained in the cells. The covering plasmid also encoded the URA3 marker, which allows for positive selection (growth on medium lacking uracil) and negative selection (growth on 5-fluro-orotic acid (FOA), a drug that kills Ura3-producing strains [78]). Plasmids with a different auxotrophic marker (HIS3) expressing no Msh2 (vector), MSH2 driven by the wild-type promoter, or MSH2 expressed from the cell-cycle box disrupted promoter were introduced into pol3-01 msh2Δ cells. In this plasmid shuffle experiment, growth on FOA indicated cells that could lose the covering MSH2 URA3-based plasmid because the introduced HIS3 plasmid produced sufficient levels of Msh2.
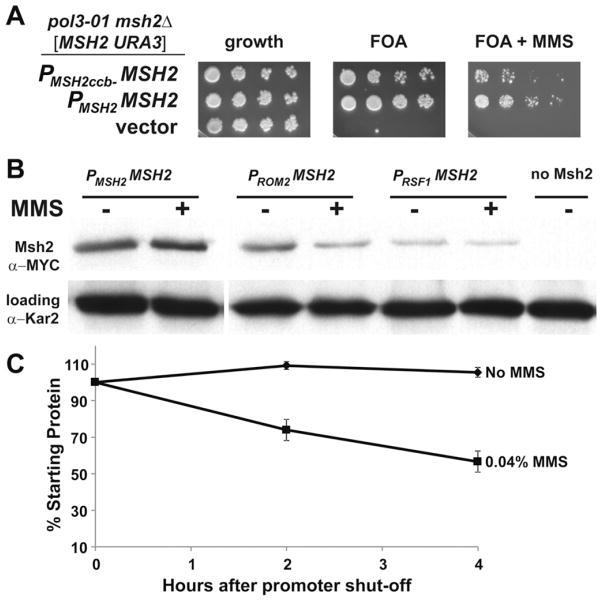
As expected, the MSH2 expressed from the wild-type promoter fully complemented the defect and the cells were FOA-resistant, in contrast to cells harboring the empty vector (Fig. 5A). When MSH2 driven by the cell-cycle box disrupted promoter was introduced, the cells were viable in the absence of MMS, but displayed a loss of viability in the presence of MMS. The data are consistent with the model that the cell-cycle regulatory elements in the MSH2 promoter are important for regulating expression upon exposure to DNA damage.
Fig. 5.
The expression of MSH2 in response to MMS is controlled at the transcriptional and post-transcriptional level. (A) Scrambling of cell-cycle elements upstream of MSH2 results reduced viability in response to MMS in a strain in which mismatch repair is essential. AGY1057, an msh2Δ pol3-01 strain kept alive by an MSH2-MYC URA3 plasmid [MSH2-MYC::KanMX6 URA3] was transformed with additional plasmids expressing: MSH2 expressed from its own promoter (PMSH2 -MSH2), MSH2 expressed from its promoter where the cell-cycle boxes are scrambled (PMSH2ccb− -MSH2), or no MSH2 (vector). Two-fold serial dilutions of exponentially growing cells were spotted onto plates containing no drug selection as a control for growth (growth), 5-FOA plates (FOA), or 5-FOA plates with 0.04% MMS (FOA + MMS). 5-FOA selects for cells that were able to lose the covering MSH2 URA3 plasmid. (B) Constitutive expression of Msh2 causes a decrease in Msh2 protein levels in response to MMS. Exponentially growing strains expressing MSH2 with a C-terminal MYC epitope tag driven from either the wild-type MSH2 promoter (PMSH2 -MSH2, strain AGY665) or the constitutive promoter upstream of ROM2 (PROM2 -MSH2, strain AGY1055), or RSF1 (PRSF1 -MSH2, strain AGY1056) were grown in the absence (−) or presence (+) of MMS for 1.5 hours and prepared for immunoblot analysis. A strain expressing an untagged MSH2 (no Msh2, strain AGY220) was included as a negative control. Protein extracts were analyzed by immunoblotting with α-MYC to detect Msh2 or with α-Kar2 as a loading control. (C) MMS increases the turnover of Msh2. A yeast msh2Δ strain (AGY75) harboring a plasmid with the GAL10 promoter fused to MSH2 (pGAL-MSH2) was used to permit controlled expression of Msh2. For the experiment, cells were grown to exponential phase in 2% galactose containing medium. The culture was split and glucose (2%) was added to each culture to repress synthesis of MSH2. One culture was also exposed to MMS (0.04%). At time intervals of 0 h, 2 h, and 4.0 h after repression of synthesis, ~3 × 107 cells were processed for immunoblotting. Protein extracts were subjected to chemiluminescence immunoblotting methods to detect Msh2 variant and a loading control. The data were normalized for growth (see Section 2) and are expressed as a percentage of the zero time point (% Starting Protein).
We tested whether the cell cycle boxes in the MSH2 promoter modulate expression in response to MMS. We measured luciferase activity with the wild-type MSH2 promoter and the cell-cycle box mutagenized MSH2 promoter fused to luciferase in the presence and absence of MMS. We found that the MMS induction is still measurable even when the cell-cycle elements are scrambled. We observed a 1.49 ± 0.04 induction for the wild-type promoter and 1.6 ± 0.2 for the cell-cycle box mutagenized promoter. These data suggest that the MMS responsiveness of the MSH2 promoter is not modulated by the cell cycle boxes. Interestingly, both protein blots and the luciferase experiments showed that the levels are 60% of wild-type levels when driven from the cell-cycle box mutagenized promoter, even in the presence of MMS (not shown). We conclude that the 40% reduction in levels is too low to support life in a pol3-01 background in the presence of mutagen MMS.
3.6. Msh2 is turned over more rapidly in response to the DNA damaging agent methyl methane sulfonate
In an attempt to further explore the importance of endogenous regulation of MSH2 and the DNA damage response, we engineered constructs with two constitutive promoters—ROM2 and RSF1—driving expression of MSH2. The promoters were selected because they do not confer cell-cycle periodicity and do not induce expression in response to DNA damage [79]. Furthermore, previous microarray analyses have revealed constitutive expression in continuous cultures under a variety of growth conditions (Maitreya Dunham, personal communication). Additionally, these promoters were selected because they produce mRNA levels of their cognate genes at levels similar (PROM2 promoter) or slightly lower than MSH2 (PRSF1 promoter), as determined in genome-wide transcription mapping experiments [80].
As expected, expressing Msh2 under the control of the two promoters described above produced similar or lower levels of Msh2 under mitotic growth when compared to expression from the endogenous promoter (Fig. 5B). However, contrary to the typical induction of Msh2 in response to MMS (~1.8 fold), Msh2 protein levels were slightly decreased when expressed from either promoter (0.7 fold for PROM2 and 0.8 fold for PRSF1). These data suggest that Msh2 is turned over at a higher rate under conditions of DNA damage.
We tested the possibility of accelerated degradation by conducting turnover experiments using the GAL10 inducible/repressible promoter to control expression of MSH2. The GAL10 promoter allows for strong expression in the presence of galactose and repression in glucose medium. Exponentially growing cells are exposed to galactose for 30 min to express wild-type Msh2. The cells were then grown in glucose to repress synthesis in the presence and absence of MMS. Samples are taken over time after repression and conventional protein immunoblotting experiments were performed. The data are consistent with an increased turnover of Msh2 in the presence of MMS (Fig. 5C).
The elevated turnover of Msh2 in response to MMS has at least two possible explanations. First, components of the ubiquitin proteasome system are known to be upregulated in response to MMS [17], and we have found that Msh2 is targeted by the ubiquitin-mediated pathway (Arlow and Gammie, submitted for publication). Thus, the enhanced turnover might be a secondary consequence of increased proteasomal activity. Alternatively, active participation in detecting damaged DNA might increase the turnover rate of Msh2, perhaps through proximity to an E3 ubiquitin ligase at chromatin or by the post-detection addition of another post-translational modification that targets Msh2 for proteasomal degradation. Employing a DNA damaging agent that does not upregulate proteasome components should provide insight into the mechanism for increased Msh2 turnover in response to DNA damage.
Supplementary Material
Acknowledgments
We are grateful to Thomas Petes for generously supplying the pSH44 plasmid and Eric Alani for providing a pol3-01 yeast strain. This research was supported by a New Jersey Commission on Cancer Research Seed Grant 10-1064-CCR-E0 awarded to A. Gammie, and New Jersey Commission on Cancer Research Summer Research Fellowships awarded to R. Tennen, H. Wijayatilake, and D. Ponzio. Finally, a National Institute of Health grant GM037739 awarded to M. D. Rose also supported this research.
Abbreviations
- uORF
upstream open reading frame
- MMS
methyl methane sulfonate
- HU
hydroxyurea
- αF
alpha factor
- WT
wild-type
- CAN
canavanine
- 5-FOA
5-fluoro-orotic acid
- MCB
MluI cell cycle box
- SCB
Swi6/4 cell cycle box
- CCB
cell-cycle box
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Jemal A, Siegel R, Xu JQ, Ward E. Cancer Statistics, 2010. Ca-a Cancer Journal for Clinicians. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Lynch HT, Lynch PM, Lanspa SJ, Snyder CL, Lynch JF, Boland CR. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 4.Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR, Petersen GM, Kinzler KW, Vogelstein B, Delachapelle A. Clues to the pathogenesis of familial colorectal-cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 5.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 6.Levinson G, Gutman GA. High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K-12. Nucleic Acids Res. 1987;15:5323–5338. doi: 10.1093/nar/15.13.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strand M, Prolla TA, Liskay RM, Petes TD. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 8.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen LA, Nystrom-Lahti M, Guan XY, Zhang J, Meltzer PS, Yu J, Kao FT, Chen DJ, Cerosaletti KM, Fournier REK, Todd S, Lewis T, Leach RJ, Naylor SL, Weissenbach J, Mecklin JP, Jarvinen H, Petersen GM, Hamilton SR, Green J, Jass J, Watson P, Lynch HT, Trent JM, Delachapelle A, Kinzler KW, Vogelstein B. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 9.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 10.Parsons R, Li GM, Longley MJ, Fang WH, Papadopoulos N, Jen J, de la Chapelle A, Kinzler KW, Vogelstein B, Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 11.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazzaro F, Giannattasio M, Puddu F, Granata M, Pellicioli A, Plevani P, Muzi-Falconi M. Checkpoint mechanisms at the intersection between DNA damage and repair. DNA Repair. 2009;8:1055–1067. doi: 10.1016/j.dnarep.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 14.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kramer W, Fartmann B, Ringbeck EC. Transcription of mutS and mutL-homologous genes in Saccharomyces cerevisiae during the cell cycle. Mol Gen Genet. 1996;252:275–283. doi: 10.1007/BF02173773. [DOI] [PubMed] [Google Scholar]
- 16.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci USA. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lotito L, Russo A, Bueno S, Chillemi G, Fogli MV, Capranico G. A specific transcriptional response of yeast cells to camptothecin dependent on the Swi4 and Mbp1 factors. Eur J Pharmacol. 2009;603:29–36. doi: 10.1016/j.ejphar.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Polager S, Kalma Y, Berkovich E, Ginsberg D. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene. 2002;21:437–446. doi: 10.1038/sj.onc.1205102. [DOI] [PubMed] [Google Scholar]
- 21.Youn CK, Cho HJ, Kim SH, Kim HB, Kim MH, Chang IY, Lee JS, Chung MH, Hahm KS, You HJ. Bcl-2 expression suppresses mismatch repair activity through inhibition of E2F transcriptional activity. Nat Cell Biol. 2005;7:U137–U148. doi: 10.1038/ncb1215. [DOI] [PubMed] [Google Scholar]
- 22.Chang IY, Jin M, Yoon SP, Youn CK, Yoon Y, Moon SP, Hyun JW, Jun JY, You HJ. Senescence-dependent MutS alpha dysfunction attenuates mismatch repair. Mol Cancer Res. 2008;6:978–989. doi: 10.1158/1541-7786.MCR-07-0380. [DOI] [PubMed] [Google Scholar]
- 23.Iwanaga R, Komori H, Ohtani K. Differential regulation of expression of the mammalian DNA repair genes by growth stimulation. Oncogene. 2004;23:8581–8590. doi: 10.1038/sj.onc.1207976. [DOI] [PubMed] [Google Scholar]
- 24.Meyers M, Theodosiou M, Acharya S, Odegaard E, Wilson T, Lewis JE, Davis TW, VanPatten CW, Fishel R, Boothman DA. Cell cycle regulation of the human DNA mismatch repair genes hMSH2, hMLH1, and hPMS2. Cancer Res. 1997;57:206–208. [PubMed] [Google Scholar]
- 25.Schroering AG, Edelbrock MA, Richards TJ, Williams KJ. The cell cycle and DNA mismatch repair. Exp Cell Res. 2007;313:292–304. doi: 10.1016/j.yexcr.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 26.Nagasaka T, Rhees J, Kloor M, Gebert J, Naomoto Y, Boland CR, Goel A. Somatic hypermethylation of MSH2 is a frequent event in Lynch Syndrome colorectal cancers. Cancer Res. 2010;70:3098–3108. doi: 10.1158/0008-5472.CAN-09-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ausubel FM, Brent R, Kingston RE, Moore DD, Scidman JG, Smith RA, Struhl K. Current Protocols in Molecular Biology. John Wiley & Sons; New York: 1994. [Google Scholar]
- 28.Kow YW, Bao G, Reeves JW, Jinks-Robertson S, Crouse GF. Oligonucleotide transformation of yeast reveals mismatch repair complexed to be differentially active on DNA replication strands. Proc Natl Acad Sci USA. 2007;104:11352–11357. doi: 10.1073/pnas.0704695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burke D, Dawson D, Stearns T Cold Spring Harbor Laboratory. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. 2000. Cold Spring Harbor Laboratory Press; Plainview, NY: 2000. [Google Scholar]
- 30.Hoffman CS, Winston F. A 10-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 31.Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 35.Longtine MS, McKenzie A, Demarini DJ, 3rd, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 36.Gammie AE, Erdeniz N, Beaver J, Devlin B, Nanji A, Rose MD. Functional characterization of pathogenic human MSH2 missense mutations in Saccharomyces cerevisiae. Genetics. 2007;177:707–721. doi: 10.1534/genetics.107.071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henderson ST, Petes TD. Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2749–2757. doi: 10.1128/mcb.12.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 39.Reenan RA, Kolodner RD. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vieites JM, Navarrogarcia F, Perezdiaz R, Pla J, Nombela C. Expression and in vivo determination of firefly luciferase as gene reporter in Saccharomyces cerevisiae. Yeast. 1994;10:1321–1327. doi: 10.1002/yea.320101009. [DOI] [PubMed] [Google Scholar]
- 41.Pringle JR, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 42.Ohashi A, Gibson J, Gregor I, Schatz G. Import of proteins into mitochondria. The precursor of cytochrome c1 is processed in two steps, one of them heme-dependent. J Biol Chem. 1982;257:13042–13047. [PubMed] [Google Scholar]
- 43.Ostapenko D, Solomon MJ. Budding yeast CTDK-I is required for DNA damage-induced transcription. Eukaryot Cell. 2003;2:274–283. doi: 10.1128/EC.2.2.274-283.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jelinsky SA, Estep P, Church GM, Samson LD. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol Cell Biol. 2000;20:8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts CJ, Nelson B, Marton MJ, Stoughton R, Meyer MR, Bennett HA, He YDD, Dai HY, Walker WL, Hughes TR, Tyers M, Boone C, Friend SH. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 46.Qiu P. Recent advances in computational promoter analysis in understanding the transcriptional regulatory network. Biochem Biophys Res Commun. 2003;309:495–501. doi: 10.1016/j.bbrc.2003.08.052. [DOI] [PubMed] [Google Scholar]
- 47.Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 48.Cliften PF, Hillier LW, Fulton L, Graves T, Miner T, Gish WR, Waterston RH, Johnston M. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 2001;11:1175–1186. doi: 10.1101/gr.182901. [DOI] [PubMed] [Google Scholar]
- 49.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 50.Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, Botstein D. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387:67–73. [PMC free article] [PubMed] [Google Scholar]
- 51.Bahler J. Cell-cycle control of gene expression in budding and fission yeast. Annu Rev Genet. 2005;39:69–94. doi: 10.1146/annurev.genet.39.110304.095808. [DOI] [PubMed] [Google Scholar]
- 52.Losi L, Di Gregorio C, Pedroni M, Ponti G, Roncucci L, Scarselli A, Genuardi M, Baglioni S, Marino M, Rossi G, Benatti P, Maffei S, Menigatti M, Roncari B, de Leon MP. Molecular genetic alterations and clinical features in early-onset colorectal carcinomas and their role for the recognition of hereditary cancer syndromes. Am J Gastroenterol. 2005;100:2280–2287. doi: 10.1111/j.1572-0241.2005.00223.x. [DOI] [PubMed] [Google Scholar]
- 53.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hood HM, Neafsey DE, Galagan J, Sachs MS. Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu Rev Micro-biol. 2009;63:385–409. doi: 10.1146/annurev.micro.62.081307.162835. [DOI] [PubMed] [Google Scholar]
- 55.Pickering BM, Willis AE. The implications of structured 5′ untranslated regions on translation and disease. Semin Cell Dev Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 56.Polymenis M, Schmidt EV. Coupling of cell division to cell growth by translational control of the G(1) cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeon S, Kim J. Upstream open reading frames regulate the cell cycle-dependent expression of the RNA helicase Rok1 in Saccharomyces cerevisiae. FEBS Lett. 2010;584:4593–4598. doi: 10.1016/j.febslet.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 58.Churbanov A, Rogozin IB, Babenko VN, Ali H, Koonin EV. Evolutionary conservation suggests a regulatory function of AUG triplets in 5′-UTRs of eukaryotic genes. Nucleic Acids Res. 2005;33:5512–5520. doi: 10.1093/nar/gki847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7:S12.1–S12.14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S-phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- 61.Bean JM, Siggia ED, Cross FR. High functional overlap between MluI cell-cycle box binding factor and Swi4/6 cell-cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics. 2005;171:49–61. doi: 10.1534/genetics.105.044560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andrews BJ, Herskowitz I. The yeast Swi4 protein contains a motif present in developmental regulators and is part of a complex involved in cell-cycle-dependent transcription. Nature. 1989;342:830–833. doi: 10.1038/342830a0. [DOI] [PubMed] [Google Scholar]
- 63.Takahata S, Yu YX, Stillman DJ. The E2F functional analogue SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters. EMBO J. 2009;28:3378–3389. doi: 10.1038/emboj.2009.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poznic M. Retinoblastoma protein: a central processing unit. Journal of Biosciences. 2009;34:305–312. doi: 10.1007/s12038-009-0034-2. [DOI] [PubMed] [Google Scholar]
- 65.Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9:738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- 66.Bean JM, Siggia ED, Cross FR. High functional overlap between MBF and SBF in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics. 2005;171:49–61. doi: 10.1534/genetics.105.044560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ho Y, Mason S, Kobayashi R, Hoekstra M, Andrews B. Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:581–586. doi: 10.1073/pnas.94.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Workman CT, Mak HC, McCuine S, Tagne JB, Agarwal M, Ozier O, Begley TJ, Samson LD, Ideker T. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yeang CH, Mak HC, McCuine S, Workman C, Jaakkola T, Ideker T. Validation and refinement of gene-regulatory pathways on a network of physical interactions. Genome Biol. 2005;6:R62.1–R62.10. doi: 10.1186/gb-2005-6-7-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Argueso JL, Smith D, Yi J, Waase M, Sarin S, Alani E. Analysis of conditional mutations in the Saccharomyces cerevisiae MLH1 gene in mismatch repair and in meiotic crossing over. Genetics. 2002;160:909–921. doi: 10.1093/genetics/160.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Greene CN, Jinks-Robertson S. Spontaneous frameshift mutations in Sac-charomyces cerevisiae: accumulation during DNA replication and removal by proofreading and mismatch repair activities. Genetics. 2001;159:65–75. doi: 10.1093/genetics/159.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morrison A, Sugino A. The 3′-->5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 75.Sokolsky T, Alani E. EXO1 and MSH6 are high-copy suppressors of conditional mutations in the MSH2 mismatch repair gene of Saccharomyces cerevisiae. Genetics. 2000;155:589–599. doi: 10.1093/genetics/155.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tran HT, Gordenin DA, Resnick MA. The 3′–>5′ exonucleases of DNA polymerases delta and epsilon and the 5′–>3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xie Y, Counter C, Alani E. Characterization of the repeat-tract instability and mutator phenotypes conferred by a Tn3 insertion in RFC1, the large subunit of the yeast clamp loader. Genetics. 1999;151:499–509. doi: 10.1093/genetics/151.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 79.Sherlock G, Hernandez-Boussard T, Kasarskis A, Binkley G, Matese JC, Dwight SS, Kaloper M, Weng S, Jin H, Ball CA, Eisen MB, Spellman PT, Brown PO, Botstein D, Cherry JM. The Stanford microarray database. Nucleic Acids Res. 2001;29:152–155. doi: 10.1093/nar/29.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tishkoff DX, Filosi N, Gaida GM, Kolodner RD. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.