Figure 1.
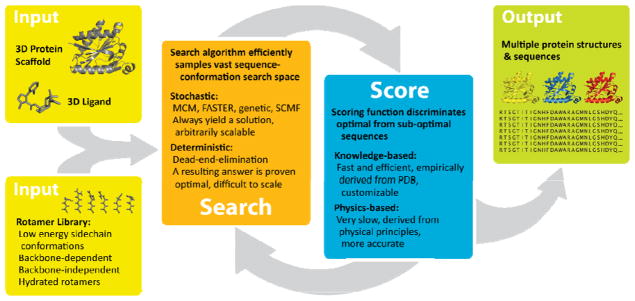
General components of an interface design algorithm, with protein and ligand structures as inputs for design. Rotamer libraries of statistically probable conformations of amino acid side chains or ligands reduce the degrees-of-freedom of the search. After multiple cycles of sequence-conformation searching followed by scoring, models are outputted that meet scoring criteria.
