Figure 3.
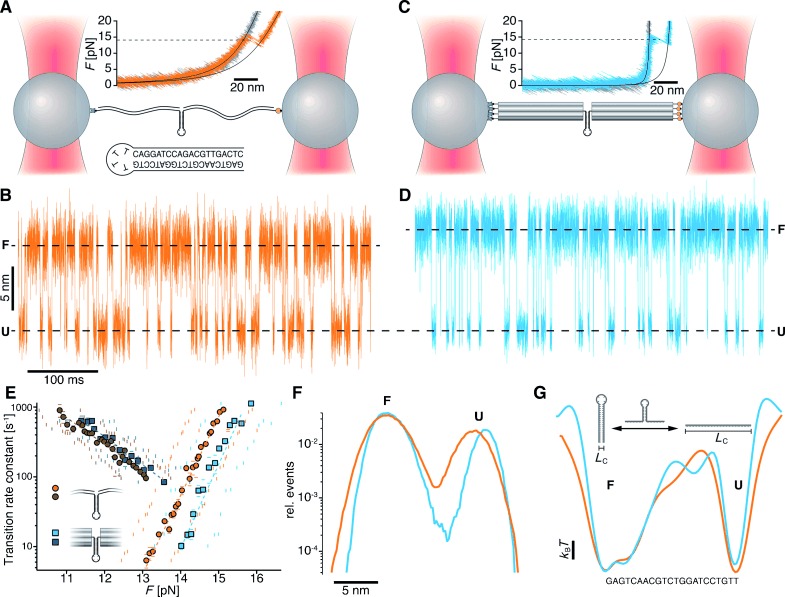
Single-molecule experiments with a stable 20 bp DNA hairpin. A,B) Conventional dsDNA linkers. A) Typical force-extension data at full measurement bandwidth. B) Typical extension data at full measurement bandwidth in constant trap distance experiments. F=folded; U=unfolded state of the hairpin. C,D) Same as in (A,B), but for experiments with ten-helix bundles. E) Force-dependent unfolding rate constants (rising branch) and refolding rate constants (falling branch) as obtained from dwell-time analysis in constant distance measurements (B,D) for different loads. The plot contains data from several molecules. Vertical and horizontal bars indicate errors of the rate constant determination. F) Histogram of the deflection states visited at a load of approximately 14 pN as measured with the conventional dsDNA (orange) versus the ten-helix bundles (cyan). G) Free-energy landscape reconstructed from (F) using deconvolution22 of the deflection statistics.
