Abstract
Inteins are rare, translated genetic parasites mainly found in bacteria and archaea, while spliceosomal introns are distinctly eukaryotic features abundant in most nuclear genomes. Using targeted metagenomics, we discovered an intein in an Atlantic population of the photosynthetic eukaryote, Bathycoccus, harbored by the essential spliceosomal protein PRP8 (processing factor 8 protein). Although previously thought exclusive to fungi, we also identified PRP8 inteins in parasitic (Capsaspora) and predatory (Salpingoeca) protists. Most new PRP8 inteins were at novel insertion sites that, surprisingly, were not in the most conserved regions of the gene. Evolutionarily, Dikarya fungal inteins at PRP8 insertion site a appeared more related to the Bathycoccus intein at a unique insertion site, than to other fungal and opisthokont inteins. Strikingly, independent analyses of Pacific and Atlantic samples revealed an intron at the same codon as the Bathycoccus PRP8 intein. The two elements are mutually exclusive and neither was found in cultured Bathycoccus or other picoprasinophyte genomes. Thus, wild Bathycoccus contain one of few non-fungal eukaryotic inteins known and a rare polymorphic intron. Our data indicate at least two Bathycoccus ecotypes exist, associated respectively with oceanic or mesotrophic environments. We hypothesize that intein propagation is facilitated by marine viruses; and, while intron gain is still poorly understood, presence of a spliceosomal intron where a locus lacks an intein raises the possibility of new, intein-primed mechanisms for intron gain. The discovery of nucleus-encoded inteins and associated sequence polymorphisms in uncultivated marine eukaryotes highlights their diversity and reveals potential sexual boundaries between populations indistinguishable by common marker genes.
Keywords: invasive elements, inteins, polymorphic introns, horizontal transfer, metagenomics, viridiplantae
Introduction
The origins and distributions of introns and inteins remain one of the greatest mysteries of molecular and evolutionary biology (Hurst and Werren 2001; Gogarten et al., 2002; Roy and Gilbert, 2006; Rogozin et al., 2012). Spliceosomal introns are distinctly eukaryotic features abundant in almost all nuclear genomes. These non-coding elements interrupt coding regions (exons) of genes and are excised from the nascent mRNA prior to translation (Roy and Gilbert, 2006; Rogozin et al., 2012). In contrast, inteins (internal protein) are much rarer genetic elements found in protein-coding genes from all three domains of life and viruses. These in-frame intervening sequences are in coding regions of genes and are translated as part of the host protein (Swithers et al., 2009). After self-catalyzed excision by the intein, the host protein flanking regions known as exteins (external protein) are ligated by a peptide bond. This intein-mediated process has been dubbed protein-splicing and maintains host protein functional integrity (Perler, 2002).
Two types of inteins are known. ‘Full inteins' possess a homing endonuclease (HE), while shorter ‘mini inteins' do not. Mini inteins are hypothesized to be remnants of ancestral full inteins from which the HE has been lost (Butler et al., 2006). HE genes encode a site-specific double-stranded DNase and are themselves mobile genetic elements (Burt and Koufopanou, 2004). HEs contribute to the spread of inteins by inducing homologous recombination at a specific cleavage site within ‘empty' alleles. HEs appear essential to intein mobility, but intein protein-splicing and HE activities are catalytically independent (Gogarten et al., 2002). Thus, even if the HE degenerates, intein splicing and host protein function remain intact. HEs are also involved in the mobility of other genetic elements, such as group I introns, but these are distinct from spliceosomal introns (which lack HEs) (Swithers et al., 2009; Rogozin et al., 2012).
Approximately 500 inteins are known from bacteria and archaea, while far fewer (∼100) have been reported in eukaryotic nuclear genomes. For the latter, inteins described thus far are almost exclusively from the supergroup Opisthokonta, specifically terrestrial microbial fungi. A few genes that are indispensable to cellular function encode most known inteins (Gogarten et al., 2002; Swithers et al., 2009). In several fungi, prp8, which encodes the essential pre-mRNA processing factor 8 protein (PRP8), contains inteins, as do key proteins VMA and GLT1 (Poulter et al., 2007). Otherwise, inteins in nucleus-encoded proteins have only been reported in the amoeba Dictyostelium discoidium and the Chlorophyceae green alga, Chlamydomonas reinhardtii (Goodwin et al., 2006). Intein evolution is not well understood and their distributions within fungal phyla are sporadic (Butler et al., 2006; Goodwin et al., 2006; Bokor et al., 2012). For instance, among prp8 genes of Cryptococcus (Basidiomycota), C. amylolentus strains have no inteins, some C. neoformans strains contain mini inteins, and C. laurentii CBS139 has a full intein (Butler et al., 2006). Such patchy taxonomic distributions have caused debate over mechanisms of intein genesis and persistence (Butler et al., 2006; Bokor et al., 2012). Inteins are primarily found at the same ‘allelic' insertion site; that is, the same insertion position in homologous genes. These sites are in regions of very high sequence conservation between both empty and invaded homologs across diverse taxa (Swithers et al., 2009).
Like inteins, spliceosomal introns can be found at the same site within gene homologs, although these positions do not appear correlated with regions of high protein sequence conservation (Gogarten et al., 2002; Swithers et al., 2009) (hereafter, unless otherwise specified, the term intron refers to spliceosomal intron). Intron gain is considered rare, and low intron numbers in an organism are often thought to reflect losses (Roy and Gilbert, 2006; Rogozin et al., 2012). While rapid intron-sequence divergence makes homologous intron prediction and comparative analyses difficult, recent population level studies are providing new insights. One of the few reports of numerous polymorphic introns shows that most were gained recently in an isolated population of Daphnia, and this crustacean had parallel gains at the same locus (Li et al., 2009). In a report on prasinophyte algae, a Pacific Micromonas had few introns while an Atlantic isolate had many more and of an unusual repetitive type (Worden et al., 2009), although whether these differences respectively reflect losses or gains remains unclear. Prasinophytes, in general, are thought to retain characteristics of the ancestor of the Viridiplantae, a kingdom containing all land plants and green algal lineages. Six closely related Prasinophyceae within the genera Micromonas, Ostreococcus and Bathycoccus have sequenced genomes (Worden et al., 2009; Vaulot et al., 2012). These picoeukaryotic (≤2 μm cell diameter) green algae belong to the order Mamiellales and are widespread in marine systems.
Here, we explored differences in prasinophyte gene complements by targeted metagenomics (Monier et al., 2012) and comparative genomics, and discovered a unique PRP8 intein in a wild Bathycoccus population. We asked whether other non-fungal taxa harbor PRP8 inteins and tested the reproducibility of our initial finding in geographically dispersed marine samples using independent molecular methods. The resulting analyses reframe phylogenetic distributions previously understood for inteins, as well as insertion site characteristics. Unexpectedly, we discovered other intervening sequences at the same Bathycoccus prp8 codon as the intein, which appear to be spliceosomal introns. Our findings introduce a complex pattern of polymorphic introns and inteins in extant taxa that reveal population structures linked to natural habitat.
Materials and methods
Sample sites, nucleic acid extraction and processing
Most samples were collected using filtration onto a 0.2 μm pore-size Supor filter after collecting seawater using Niskin bottles. DNA was extracted from these filtered seawater samples (Supplementary Table 1) and cultured Bathycoccus prasinos CCMP1898 (obtained from the National Center for Marine Algae and Microbiota) using a modification of the DNeasy kit (Qiagen, Germantown, MD, USA) as described previously (Demir-Hilton et al., 2011). The wild Bathycoccus-targeted metagenomic scaffolds were generated using multiple displacement amplification, sequencing and assembly on tropical Atlantic cells sorted away from other resident microbes based on fluorescence and scatter characteristics using flow cytometry (Monier et al., 2012).
Homing endonuclease and intein detection
Presence of protein domains/families shared between wild Bathycoccus and other Mamiellales, or unique to each, was determined by: (i) predicting open-reading frames on Bathycoccus scaffolds, with a minimum open-reading frame size of 60 amino acids; (ii) retrieving predicted proteomes in NCBI RefSeq non-redundant (nr) database or JGI for Micromonas pusilla CCMP1545 and M. sp. RCC299, Ostreococcus tauri, O. lucimarinus and O. sp. RCC809; (iii) searching these using hmmsearch, part of HMMer v3, with the Pfam-A collection (Finn et al., 2010; Eddy, 2011). Absence of the HE (Pfam model PF05203) was confirmed by negative tBLASTn results against the Mamiellales genomes using the wild Bathycoccus PRP8 full intein protein sequence (encoded on contig C1684, accession AFUW01000096). Similar searches were conducted against nr, the Salpingoeca rosetta and Capsaspora owczarzaki genomes (Nichols et al., 2012), and JGI algal genomes (Emiliania huxleyi, Fragilariopsis cylindrus, Coccomyxa C-169 and Bigelowiella natans), using wild Bathycoccus and four fungal PRP8 full intein protein sequences (NEB InBase identifiers: Ascomycota, Hca-PRP8; Basidiomycota, Cla-PRP8; Chytridiomycota, Bde-JEL423-PRP8; Mucoromycotina, Pbl-PRP8-a) as queries. Intein splicing motifs flanking the HE domain were identified using InBase data (Perler, 2002) and aligned using MUSCLE v3.8 (Edgar, 2004) with manual editing.
PCR, cloning and sequencing
Primers were designed to extein regions, so the amplicon would span the intein, using prp8 genes from wild Bathycoccus and isolate BBan7. PRP8f and PRP8r annealed to extein regions, spanning insertion sites and amplifying 200 extein nucleotides, while intein-specific primers PRP8if2 and PRP8ir were paired with extein primers to amplify part of the intein and provide overlapping products. For each DNA extract three PCRs were performed using PRP8f/PRP8r, PRP8if2/PRP8r and PRP8f/PRP8ir (Supplementary Table S1). When multiple bands were observed in prp8 PCR reactions, we cloned into pCR2.1 (Invitrogen, Grand Island, NY, USA) and performed colony PCRs (vector-specific primers M13F and M13R) to screen and select plasmids representing each product type. Plasmids were purified (QIAprep, Qiagen) according to the manufacturer's procedures. Sequencing was performed on an ABI37130xl (Applied Biosystems, Grand Island, NY, USA). In addition to prp8 gene sequence and intervening sequences recovered by the above primers, PRP8f/PRP8ir amplified a partial intein-size like band in the North Atlantic Slope and Central North Pacific samples, which were an unspecific (non-PRP8/intein) product (BLASTx hits to hypothetical bacterial genes); PRP8if2/PRP8r showed no amplification at these sites (but did at sites where PRP8 inteins were recovered). NCBI accession numbers are provided in Supplementary Table S2.
Phylogenetic analyses
For extein phylogenetic analysis, 46 PRP8 homologs were retrieved from various databases using BLASTp searches with the wild Bathycoccus PRP8 as a query (Supplementary Table S3) and aligned using MUSCLE v3.8 (after intein removal). After manual curation ambiguously aligned positions and those with >50% gaps were discarded. The maximum-likelihood tree was reconstructed using phyML v3 (Guindon and Gascuel 2003) and the LG+I+G substitution model, selected as the best fit by ProtTest v3 (Darriba et al., 2011) according to Akaike information criterion. Node support was computed using 100 bootstrap replicates.
Prior to 18S and 5.8S rRNA genes and ITS2 phylogenetic analyses, we identified the wild Bathycoccus scaffold encoding those genes (C561; accession AFUW01000141) and also PCR amplified, sequenced and deposited (JX625115) this region from B. prasinos CCMP1898 using primers 1400F and ITS055R (Marin and Melkonian 2010). Sequences were manually added to a ‘reference alignment', focused on the proposed class Mamiellophyceae (Marin and Melkonian 2010), which was then edited manually and masked. Trees were reconstructed in phyML v3 using best-fitting models (see legends).
Intein splicing domains were analyzed using conserved blocks A, B, F and G retrieved from InBase (from both prp8 and other intein-containing genes) and concatenated. Splicing motifs from the PRP8 inteins of wild Bathycoccus, the PCR sequences, C. owczarzaki and S. rosetta were added manually. The global splicing motif tree was computed using FastTree v2.1 (Price et al., 2010) in ‘slow and accurate' mode (-spr 4 -mlacc 2 -slownni) and the JTT model. Local statistical support was computed using Shimodaira-Hasegawa-like tests with 1000 replicates. The same methodology was employed for HE reconstructions based on blocks C, D, E and H. Only HEs with all four conserved blocks were used, and those of the wild Bathycoccus PRP8 inteins, PCR sequences, and C. owczarzaki were aligned manually. For both splicing and HE motif alignments, we also constructed distance-based trees using BIONJ (Gascuel 1997) and computed distances and bootstraps (100 replicates) using PHYLIP v3.69 modules PROTDIST and SEQBOOT (Felsenstein, 2005). Amino acid multiple sequence alignments of PRP8 exteins, as well as intein splicing and HE motifs, along with trees, were deposited at treebase.org (Piel et al., 2009) under project number 13909.
Residue conservation plot
Overall residue conservation was computed using the PRP8 extein tree alignment and a 10-residue sliding window with methodology and scripts as in (Swithers et al., 2009).
Results
Discovery of a full intein in an algal prp8 gene
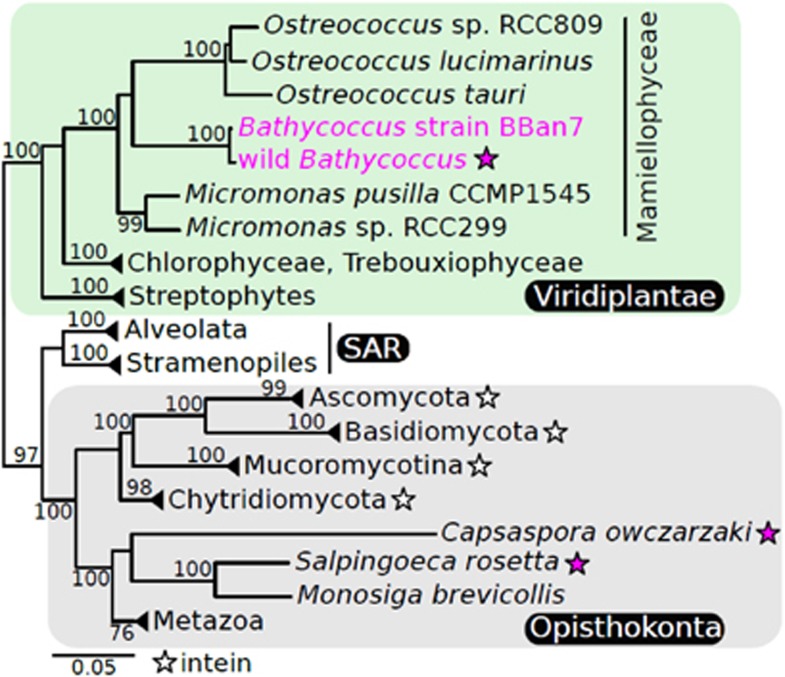
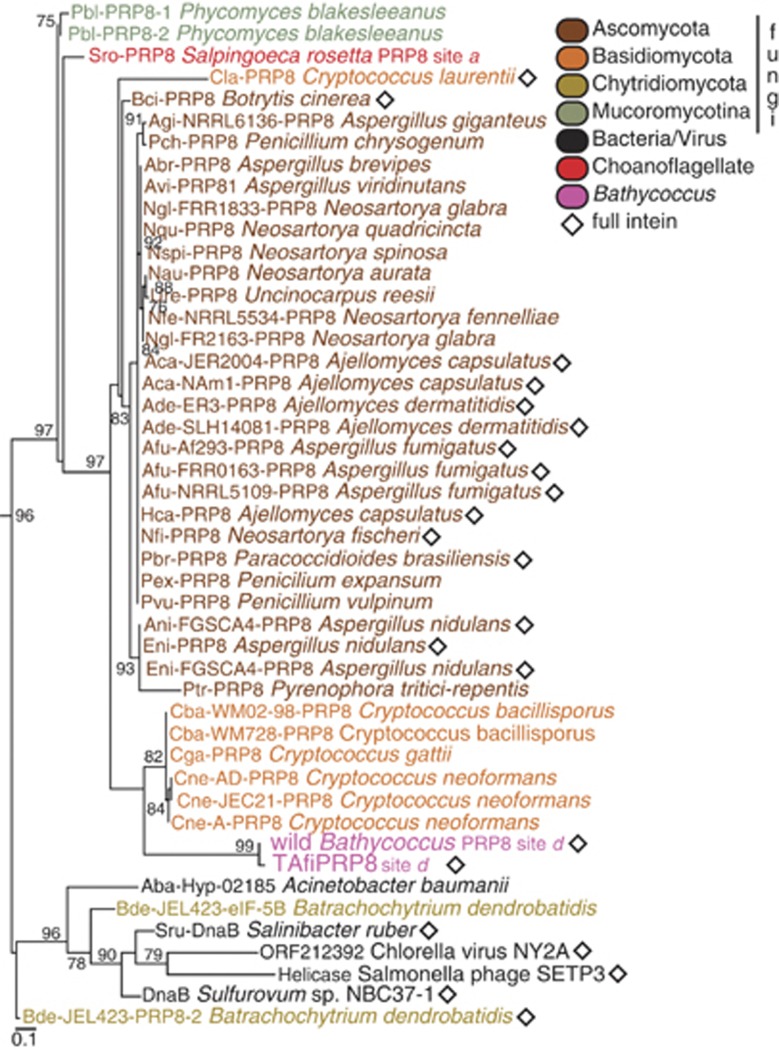
We compared the predicted proteome from a wild population of Bathycoccus to those available from other Mamiellales. The wild Bathycoccus population was collected by fluorescence activated cell sorting from the tropical Atlantic Ocean and, after multiple displacement amplification of DNA from the retrieved cells, its genome was partially sequenced (Monier et al., 2012). The wild Bathycoccus contained a putative HE domain embedded in its prp8 gene that was not found in other Mamiellales genomes. Phylogenetic analysis of corresponding protein sequences showed that the wild Bathycoccus PRP8 was closely related to that from Bathycoccus prasinos BBan7 (Figure 1) a cultured strain that has a sequenced genome (Vaulot et al., 2012).
Figure 1.
Maximum-likelihood phylogenetic analysis of PRP8 protein sequences using 1921 extein positions. Stars denote groups with previously known (black) intein-containing members and those identified herein (fuchsia). Stramenopiles, Alveolates and Rhizaria are abbreviated as SAR. The scale bar reflects substitutions per site. Bootstrap support ⩾75% is shown.
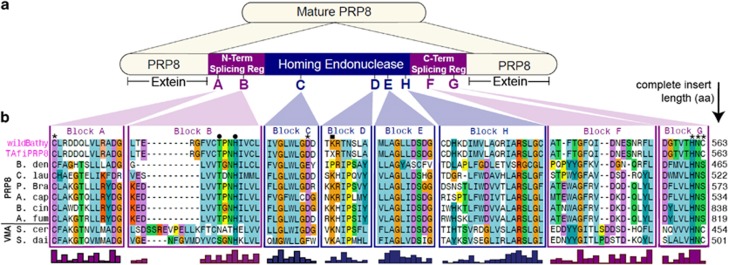
The 563 amino-acid insertion in the wild Bathycoccus PRP8 sequence includes 194 HE residues. The N- and C-termini of the inserted sequence contained all known intein splicing motif blocks (A, B, F and G) and all blocks of conserved HE residues (C, D, E and H) (Figure 2). These analyses indicated the insertion is a full intein.
Figure 2.
Intein domain architecture. (a) Schematic of extein, conserved intein-splicing motif (blocks A, B, F and G), and homing endonuclease (blocks C, D, E and H) domains. (b) Alignment of PRP8 intein splicing and HE motifs from the wild Bathycoccus targeted metagenome, environmental clone TAfiPRP8 (from unsorted Tropical Atlantic DNA) and fungal representatives from different insertion sites. Essential (asterisks) and non-essential (dots) splice junction residues and HE catalytic (asterisks) and other essential (square; lysine, K) residues are indicated. Residue ‘X' in TAfiPRP8 block D represents an ambiguous base call resulting in possible codons AAG or CAG (residue K or Q). Conserved residues are colored according to physicochemical properties (ClustalX color scheme). Histogram shows overall conservation levels; amino acids located between bracketed blocks were omitted.
Identification of additional non-fungal PRP8 inteins
As so few inteins have been observed in eukaryotes other than fungi, and none has been reported in non-fungal prp8 genes, we searched NCBI RefSeq nr and other databases using tBLASTn and BLASTp with the wild Bathycoccus PRP8 intein as a query. Prp8 homologs containing inteins were identified in two non-fungal, marine opisthokonts. Capsaspora owczarzaki had three full inteins in its prp8 gene and Salpingoeca rosetta had a 201 residue mini-intein.
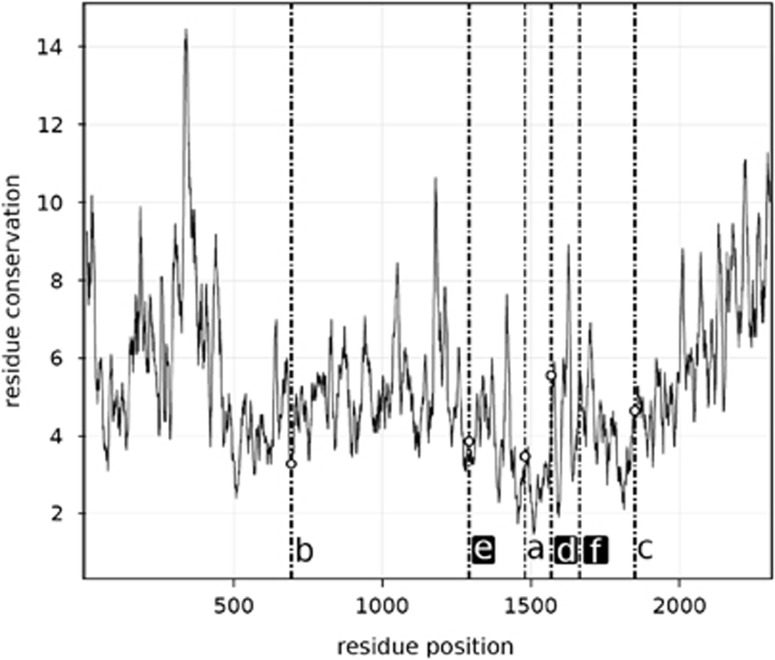
Insertion sites for most newly identified inteins were different from those of known PRP8 inteins, which are at site a, except one each from the chytrid fungi Spizellomyces punctatus (site c; Spu-PRP8) and Batrachochytrium dendrobatidis (see InBase database). The latter has two: one at site a, and another further upstream (site b; Bde-JEL423-PRP8-1). The wild Bathycoccus PRP8 intein is 87 residues downstream from site a at a new insertion site herein termed site d (Figure 3, Supplementary Figure 1). In S. rosetta the mini intein was at site a and C. owczarzaki had one intein at site b. The other two C. owczarzaki PRP8 inteins were at novel sites named here e and f. Conserved splicing motifs and essential catalytic residues were present in the newly identified PRP8 inteins (Figure 2, Supplementary Figure 1). Insertion sites ranged in amino-acid conservation levels (Figure 3).
Figure 3.
Intein insertion sites (dashed lines) along 2309 residues of PRP8 and the corresponding amino-acid conservation profile for taxa from different eukaryotic supergroups (as in Figure 1). Higher Y-axis values indicate lower conservation at that position (that is, a greater number of different amino acids). Dots show exact insertion sites and newly identified sites are in black frames.
Novel intervening sequences in Bathycoccus prp8 genes
In contrast to the wild Bathycoccus population, cultured strains tested did not contain intein sequences within the prp8 locus. PCR primers designed to flank the wild Bathycoccus insertion site amplified prp8 gene sequence from B. prasinos CCMP1898. However, no insertion was found in this strain or BBan7. Primers designed internal to the intein sequence, to avoid potential bias for empty alleles over large inserts, did not produce products from cultures, again indicating the cultured Bathycoccus strains possessed only intein-lacking prp8 alleles.
Despite this distinct difference in prp8 genetic architecture, several lines of evidence demonstrate the wild sequence is from Bathycoccus. Phylogenetic reconstruction of PRP8 exteins provided strong statistical support for their taxonomic relatedness to cultured B. prasinos (Figure 1). The 18S rRNA gene from the Bathycoccus targeted metagenome had 100% identity to those from cultures, an identity level used previously to infer all Bathycoccus belong to a single species. Phylogenetic analysis of wild and cultured Bathycoccus sequences showed no (18S rRNA gene), or relatively low (ITS2-5.8S) divergence and formed a supported clade nested within Mamiellales (Supplementary Figures 2 and 3), as commonly reported (Marin and Melkonian, 2010; Worden, 2006).
As we found only empty prp8 alleles in the cultured Bathycoccus strains, we turned to genetic material from the ocean to try to reproduce our initial finding for the wild population. The Bathycoccus prp8 PCR primers were applied to filtered samples (first from the same tropical Atlantic site as the targeted metagenome) and confirmed the presence of an intein in Bathycoccus prp8 genes in nature. This independent result ruled out the possibilities that technical issues (for example, chimeras) influenced our discovery of a wild Bathycoccus PRP8 full intein, including issues potentially arising from using multiple displacement amplification or low scaffold fold coverage (4±2 (s.d.)).
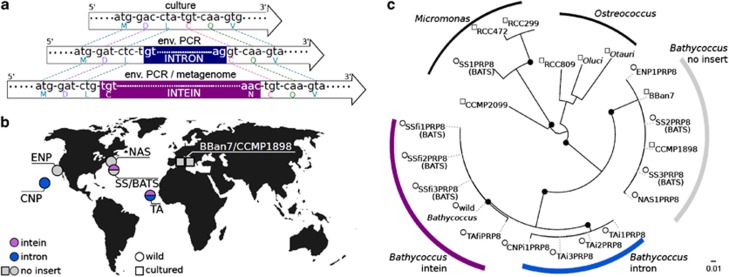
Two types of environmental Bathycoccus sequences were discovered in cloned tropical Atlantic (TA) prp8 PCR amplicons, both bearing insertions (Figure 4a, Supplementary Table 2). Prp8 coding regions of all clones had 100% amino-acid identity to wild and cultured Bathycoccus, except TAfiPRP8 which had one different residue. The first type contains a full intein (clone TAfiPRP8, fi for full intein, Figure 2b). We hypothesize the second type contains an intron; it had a small 135 bp insert (for example, clone TAi1PRP8, i for intron). The insert was at the same codon as the intein, but at phase 1 (in which the donor site is between the first and second codon positions). The 5′ and 3′ regions flanking the first and second prp8 intervening sequence types had 99% and 98% nucleotide identities, respectively, to the wild Bathycoccus population and 91–95% to B. prasinos CCMP1898 and BBan7, but much lower identity to Ostreococcus (80–84%) and Micromonas species (for example, RCC299, 75–78%). Thus, both environmental PCR product types appeared to be from Bathycoccus, not other Mamiellales.
Figure 4.
PRP8 polymorphic intervening sequences in Bathycoccus. (a) Schematic of Bathycoccus intervening and insert-less sequence types. (b) Geographic locations of polymorphic Bathycoccus PRP8 sequences. (c) Phylogenetic relationships among PRP8 coding sequences (nucleotides) that flank intein insertion site d of wild Bathycoccus. Phylogenetic grouping is driven by variations at the third-codon position and this tree should not be used for evolutionary distances as these positions may be saturated. Node support ⩾75% is shown (black dots).
The four TA clones with small inserts were identical in prp8 coding regions (nucleotide level). Their intervening sequences had poly-pyrimidine stretches characteristic of introns. Overall, the putative introns had 49% T, but only 22%, 16%, 13%, C, G and A, respectively. In contrast, flanking sequences had a more balanced composition with 27% T, and the overall targeted (meta)genome sequence being 48% G+C (Monier et al., 2012). We were unable to identify branch points, which are also poorly conserved in Ostreococcus (Irimia and Roy, 2008). Canonical spliceosomal intron splice sites (donor: GT, acceptor AG) were present (Figure 4a). Two clones were identical (deposited as TAi3PRP8), while the intronic sequences in TAi1PRP8 and TAi2PRP8 were 98% identical and each had 97% identity to TAi3PRP8 (Supplementary Figure 4). No similar sequences were found in Genbank and no cultured Mamiellales sequenced to date contained an intron at this site.
Environmental distribution of intervening sequences
With initial results revealing three different types of Bathycoccus prp8 gene structure, we further investigated environmental distributions. In metagenomes, we recovered two prp8 types, but only one at any particular geographical location. Six reads corresponding to the intein (for example, 98% nucleotide identity, E-value 1 × 10−124, CAMERA ID: BATS_Read_05201288) were found at the Bermuda Atlantic Time Series Site (BATS) in the Sargasso Sea, using BLASTn searches with the wild Bathycoccus prp8 intein. The intron-containing type was present in a metagenome from station ALOHA in the central North Pacific Ocean (CNP; HF_Read_03445078). The first 142 nt of this 454 read belonged to prp8 (100% identical to TAi3PRP8); the remaining 132 nt had 97% identity to the TA intron sequence, ending just before the acceptor site (Supplementary Figure 4).
PCR on additional marine samples recovered the three prp8 types, but not all together in one sample (Figure 4b). Insert-less and intein-containing prp8 versions were found at BATS (Figure 4b). On the North Atlantic slope and eastern North Pacific only the insert-less type was detected, while the other Pacific site (ALOHA) had the intron-containing type. In addition to strong bands represented by the above sequences, faint bands could not be cloned from two filtered samples, BATS and TA. Products were similar in size to intron-containing and no-insert amplicons, respectively. These bands, especially the no-insert size, may represent products from other algae. Our primers amplified Micromonas (Figure 4c) and non-prasinophyte algal prp8 genes (for example, Pelagomonas), none of which contained an insert. Overall, environmental prp8 sequence identities followed patterns established between TA and insert-less culture sequences, with insert-bearing sequences having very high identity in gene coding regions. Phylogenetic reconstruction using prp8 extein/exon coding regions supported this observation (Figure 4c).
Relationships among PRP8 inteins and HEs across lineages
We performed phylogenetic analyses on intein-splicing and HE motif blocks from bacteria, archaea, eukaryotes and viruses. Block residues were used rather than other regions, which were highly divergent. This approach allowed us to explore general phylogenetic trends, but did not have the resolution to assess finer scale relationships due to the limited number of positions analyzed. Site a PRP8 inteins grouped together in a supported clade that also included wild Bathycoccus inteins when analyzed by approximate maximum-likelihood (Figure 5), but not distance methods (Supplementary Figure 6). Other non-site a PRP8 inteins were placed elsewhere, specifically site b and c chytrid inteins and C. owczarzaki inteins (Supplementary Figure 5). A clade related to the primary PRP8 group contained full inteins from a virus (PbCV-NY2A) that infects the eukaryotic green alga Chlorella (in a protein of unknown function), a phage SETP3 protein, B. dendrobatidis eIF-5B, and bacterial proteins (Figure 5, Supplementary Figure 6), but was only supported by approximate maximum-likelihood methods (Figure 5).
Figure 5.
Phylogenetic relationships among intein splicing motifs from Bathycoccus (both metagenomic and PCR-based), the choanoflagellate S. rosetta, fungi, and the nearest outside group (inteins from bacterial and viral genes) are shown in this subtree derived from an analysis of 453 inteins (Supplementary Figure 5). Approximate maximum-likelihood inference was used on 51 amino-acid positions, representing conserved blocks A, B, F and G. NEB InBase identifiers precede species names. Node support is indicated where ⩾75% and substitutions per site (scale bar) are shown. PRP8 inteins are at site a unless noted otherwise.
The newly identified HEs belonged to the LAGLIDADG family along with all other eukaryotic full intein HEs (Gogarten et al., 2002). HEs from Bathycoccus and two C. owczarzaki PRP8 inteins branched together, in the eukaryotic HE region of the tree (Supplementary Figure 7), but separate from other PRP8 HEs in an unsupported position (Supplementary Figures 8 and 9). Those of C. owczarzaki site f and S. rosetta were even more divergent (Supplementary Figure 7).
Discussion
We used targeted metagenomics to retrieve genomic information from a wild population of a marine green alga (Cuvelier et al., 2010; Monier et al., 2012). An intein was discovered in wild Bathycoccus that is absent from sequenced algal genomes, including cultured Bathycoccus. The Bathycoccus intein is in the largest protein of the spliceosomal machinery, PRP8 (Dlakic and Mushegian, 2012). It represents the first PRP8 intein described in the Plantae, or indeed outside of fungi, and is in the size range of fungal PRP8 full inteins (642±138 (s.d.)). We also identified inteins in available sequences from the marine protists C. owczarzaki, a parasite that forms an outgroup to metazoans and choanoflagellates, and S. rosetta, a choanoflagellate (Ruiz-Trillo et al., 2008; Nichols et al., 2012). These are the first non-fungal opisthokont PRP8 inteins reported. Until now, only RNA polymerase sub-units were known to have inteins spanning two eukaryotic supergroups, specifically in B. dendrobatidis (Opisthokonta; subunit II, RPB2 and III, RPC2), and one sub-unit in C. reinhardtii (Plantae; RPB2) and D. discoideum (Amoebozoa; RPC2) (Goodwin et al., 2006). Here, results from independent samples and methods confirmed the Bathycoccus targeted metagenome assembly containing the intein and led to the discovery of multiple types of intervening sequences. The results reveal plasticity in genetic elements and gene architecture that appears to be connected to ecological distributions.
PRP8 inteins and patchy phylogenetic distributions
The new inteins contain motifs indicative of intact self-splicing capabilities, as expected given that failed splicing would prevent synthesis of a functional, mature PRP8 protein. The Bathycoccus full intein, and two of the three in C. owczarzaki, were at unique insertion sites, doubling the number of recognized PRP8 intein sites (Figure 3, Supplementary Figure 1); 39 of the 41 known fungal PRP8 inteins are at site a (see InBase database). Additionally, Bathycoccus and C. owczarzaki inteins appear capable of homing since they contain HE blocks and residues required for homing displacement.
One could expect to see the prp8 gene harboring genetic parasites, even though PRP8 inteins have previously only been observed in fungi. PRP8 is essential for all intron-containing eukaryotes, making mutation likely to be deleterious, and loss fatal. It has high amino-acid identity, >60% between unicellular (fungi) and multicellular (mammals) opisthokonts as well as between fungi and plantae (Dlakic and Mushegian, 2012), which may widen species boundaries of homing displacements if coding sequence identity is even higher in certain regions (for example, at potential nucleotide recognition sites). However, we found PRP8 intein insertion sites were not at the most conserved residues, and conservation levels were lower (Figure 3) than for sites in other genes; RFC, CDC21 (archaeal proteins) and VMA (yeast and archaea) have intein insert sites with values between 2–4 that are at the most conserved positions (Swithers et al., 2009). As conservation estimates are influenced by taxon sampling, one might expect higher site conservation for PRP8, which is solely eukaryotic, than VMA across two domains of life. Interestingly, PRP8 inteins are within (sites a, c to f) or close to (site b, position 735) the core region where protein-RNA contacts occur, between positions 770 and 2173 (Dlakic and Mushegian, 2012). This core region has similarities to the catalytic domain of reverse transcriptases encoded by retro-elements in archaea and bacteria, and has thus been hypothesized to be retro-element derived (Dlakic and Mushegian, 2012). In this hypothesis, the ancestral version of prp8 was a parasitic retro-element recruited early on to perform a spliceosomal role by the emerging eukaryotic cellular machinery. If so, it is intriguing that the prp8 gene itself was invaded by genetic parasites: inteins.
Allelic inteins (for example, PRP8 inteins at site a) typically have higher sequence similarity with each other than non-allelic inteins, presumably a consequence of homing displacement (Gogarten et al., 2002; Ogata et al., 2005; Poulter et al., 2007). Here, phylogenetic analysis of PRP8 intein splicing motifs indicated the Bathycoccus intein is more closely related to those in Dikarya (Ascomycota and Basidiomycota, all site a) than in C. owczarzaki, S. rosetta, Chytrid or Mucoromycotina fungi, at least based on the limited number of positions analyzed (Figure 5, Supplementary Figures 5 and 6). This was surprising because PRP8 inteins from the latter two (Bde-JEL423-PRP8-2 and Pbl-PRP8-a) are at site a, but the Bathycoccus intein is not. In addition, the C. owczarzaki PRP8 inteins at site b and newly identified site e, were quite divergent and most related to each other (Supplementary Figures 5 and 6).
The puzzling phylogenetic distributions of the newly identified inteins can be reconciled by different evolutionary scenarios such as spread by horizontal transfer (HT) and/or independent losses in distinct lineages. Of models developed to explain intein presence/absence patterns in homologous fungal genes, a prevalent one involves invasion, spread via meiosis, fixation, degeneration, loss and sometimes reinvasion; once all sites in a population are occupied by the intein, there is no longer selection for HE (Burt and Koufopanou, 2004). Thus, in this model, HE persistence is thought to require HT to another species before all alleles are occupied (Gogarten et al., 2002; Burt and Koufopanou, 2004). However, modeling of HE behavior and simulations for sexual populations indicate HT is not needed for maintenance of HEs (Yahara et al., 2009), although here effective population sizes, frequency of meiosis and cost of pseudogenes become key. Fixation does not seem to occur in some fungi, hence some alternative models do not invoke HT but instead suggest intein-occupied and empty alleles remain in equilibrium (Butler et al., 2006; Yahara et al., 2009; Bokor et al., 2012).
Prior to finding additional environmental and opisthokont PRP8 inteins, HT of the intein to—or from—Bathycoccus seemed the most straightforward explanation for sporadic distributions. The HT hypothesis minimized the evolutionary events needed to obtain the observed presence/absence patterns, although clearly taxon undersampling can cause spurious phylogenetic patterns (and should therefore be considered). The patchy phylogenetic distribution of PRP8 inteins, and generally of all inteins, could also be explained by more ancient origins and loss of these genetic inserts in distinct lineages over time (Pietrokovski 2001). Indeed, the presence of PRP8 inteins in C. owczarzaki and S. rosetta supports the idea that an ancestral opisthokont possessed this feature. Still, the S. rosetta site a mini-intein appeared more closely related to fungal site a inteins than inteins of C. owczarzaki (Figure 5, Supplementary Figures 5 and 6), its close relative according to organism (Ruiz-Trillo et al., 2008) and extein phylogenies (Figure 1). This suggests long divergence times or different origins where, if ancestral, C. owczarzaki may have lost the site a allelic intein. Additional sequencing may reveal broader distributions and a source as yet unknown.
Intervening sequences, suggestive of introns
Smaller intervening sequences discovered in natural Bathycoccus had features typical of introns but not of inteins or HEs (Figure 4, Supplementary Figure 4). None of the six cultured, genome-sequenced Mamiellales have introns in the vicinity of PRP8 intein site d. While intein distributions are often ‘polymorphic', depending on whether all alleles have been colonized, polymorphic introns have rarely been reported (Li et al., 2009). Although the putative prp8 introns had canonical splice sites, and were inserted at the same site d codon, they were phase 1 rather than between codons (phase 0, Figure 4a). Most eukaryotic introns are phase 0; phase 1 and 2 introns are thought to have more deleterious effects related to faulty splicing and intron sliding (Lynch, 2002). Thus, phase 1 positioning in prp8 may indicate these introns are not fixed in the population and were gained relatively recently.
Origins and indicators of sex and diversity
Previous environmental 18S rRNA studies suggest that although Bathycoccus is widespread, it is very homogeneous in terms of genetic diversity (Marin and Melkonian, 2010). Likewise, 18S rRNA genes analyzed herein showed no divergence (Supplementary Figure 2). Thus, what initially seemed to be a patchy distribution of inteins in Bathycoccus prp8 genes was somewhat surprising, but might have simply indicated that insert-less prp8 sequences represented alleles within a species that had yet to be colonized. However, significant sequence variations were observed between prp8 without site d insertions (Mediterranean cultures) versus those with inteins (TA, BATS) and introns (TA, CNP) (Figure 4). Insert-less prp8 were distinct from the insert-bearing types and had considerable third codon variation from them (Figures 4a and c). In contrast, intein- and intron-containing prp8 sequences grouped together and had only a single third-codon nucleotide difference between them. Moreover, insert-less isolates had no divergence from each other based on the ITS2-5.8S segment (which is less constrained than 18S rDNA) but formed a separate (sister) branch to intein-containing wild Bathycoccus (Supplementary Figure 3). Thus, an alternative hypothesis is that the empty versus colonized prp8 types represent sexually incompatible populations.
The different Bathycoccus prp8 types also appear to be associated with specific environmental settings. In our limited survey, intein/intron containing Bathycoccus prp8 sequences were recovered from more nutrient poor, open-ocean waters (See also Monier et al., 2012) and were only observed in wild populations. For example, in CNP and TA populations, prp8 was occupied by either site d introns or inteins at the time sampled. Insert-less types were found at sites or times of higher nutrients, such as the Gulfs of Lion (France, BBan7) and Naples (Italy, CCMP1898) and BATS in spring. Furthermore, the mesotrophic Eastern North Pacific (ENP) and North Atlantic Slope (NAS) sites showed only insert-less prp8 (Figure 4). We also identified the insert-less prp8 in a Bathycoccus targeted metagenome from Chilean coastal waters (Vaulot et al., 2012), which had highest identity to the Mediterranean cultures. This indicates that mesotrophic Bathycoccus populations may generally lack intervening sequences and maybe less diverse than those in more nutrient-poor settings. Although negative PCR results could occur when an allele is present at low abundance (as opposed to being absent), they serve as stronger evidence of allele absence than negative results from metagenomes. The latter are subject to interpretation issues arising from the number and diversity of templates as well as differential organism abundances. Here, open-ocean Bathycoccus populations appeared to be more diverse than those in mesotrophic settings, a complexity seemingly underrepresented by molecular surveys of 18S rDNA sequences and studies with overrepresentation of coastal samples. Still, although there are differences in environmental distributions observed here, both insert-less and -containing sequences were observed in the same BATS sample, so that if these populations were sexually compatible, intein exchange should have been possible.
Together our results indicate that at least two putative ecotypes exist (one associated with more mesotrophic environments and the other more open-ocean environments) and could well be sexually isolated. Thus, the polymorphic alleles could be used to trace sexual reproduction/isolation and dispersion across marine provinces with higher resolution data sets. With respect to the insertions themselves, at least three evolutionary scenarios could have led to the observed polymorphisms: (i) an ancestral intron might have been replaced by the intein (and possibly lost in other populations); (ii) once colonized by an intein, site d may have been an entry gate for other elements (such as introns) through recombination, or, (iii) initial colonization by an intein could have resulted in degeneration into an intron-like element. The simplest explanation is that the putative introns were gained through mechanisms like (ii) or (iii), given that other prasinophyte and some Bathycoccus prp8 genes do not contain similar intervening sequences. Without reference to introns, a model has been proposed in which inteins do not become fixed within a population but eventually turn into polymorphic elements (Butler et al., 2006). This model appears consistent with our observations although more extensive population genetic studies are needed for validation, and questions remain regarding the fitness consequences of intervening sequences—and what other levels of genomic sequence variation they represent.
Ecology of invasive element transmission and propagation
Independent from sex, invasive elements can spread by physical encounter of HE/genetic material from different cells. At low abundance, or in dilute environments like the ocean, even when effective population sizes are large, encounter rates are potentially lower than in complex terrestrial communities associated with living and detrital material. In dilute conditions, an organism acting as a hub for exchange of genetic material (Skippington and Ragan, 2011) may facilitate successful homing (or exchange). Amoebae purportedly serve as such hubs for horizontal gene transfer across bacteria (Ogata et al., 2006; Moliner et al., 2010), and, interestingly, the amoeba D. discoideum has an intein. C. owczarzaki interacts with sporocysts of worms that co-reside in the aquatic snail it parasitizes (Owczarzak et al., 1980); its host snail may be a hub for multiple microbes. S. rosetta has complex microbial interactions and could itself serve as such a hub.
Ocean predatory and parasitic protists may also facilitate spread of invasive elements. However, transmission of inteins between distant organisms and colonization requires successful homing—the recipient genome must have compatible recognition sites. Although some inteins are in highly conserved regions (Swithers et al., 2009), distantly related eukaryotes may not have sufficient conservation of specific (HE) target nucleotide motifs. We propose that viruses facilitate cross-species transmission of inteins by spawning stochastically modified HE recognition sites, generating genetic diversity of these parasitic selfish elements. Full inteins are present in various viral genomes, including those infecting insects (Pietrokovski, 2001), amoebae (Ogata et al., 2005), and two heterokonts (Nagasaki et al., 2005; Goodwin et al., 2006). Viral DNA polymerases are more error-prone than their cellular counterparts, leading to random mutations caused by lower-fidelity DNA synthesis (Sanjuan et al., 2010). Therefore, it seems possible that PRP8 intein occupation of multiple different insertion sites in fungi, Bathycoccus and C. owczarzaki (Supplementary Figure 1), or their ancestors, was facilitated by viral alteration of the intein-HE homing sequence.
Marine phycodnavirus DNA polymerase genes in nature, including prasinoviruses, have inteins (Nagasaki et al., 2005; Culley et al., 2009). Our observation of a full intein in a green algal phycodnavirus (PbCV-NY2A) related to PRP8 inteins (Figure 5), along with presence of inteins in prasinoviruses, make it tempting to speculate that the Bathycoccus PRP8 intein, or that of an ancestor, was introduced by a virus to the host. While potential hub organisms like predatory protists may facilitate exchanges between co-located microbes, viruses alter encounter rates dramatically because of their high abundance relative to host cells. If viruses randomly alter HE recognition sites, and serve as intein carriers, they could play a significant role in transmission to new loci or empty alleles in distant hosts.
Conclusions
Our findings highlight the diversity of uncultivated microbial eukaryotes at the level of gene architecture. The variety of polymorphic introns and inteins discovered here has been missed by culture studies, and likely reflects important, understudied aspects of ecological fitness and differentiation. These elements highlight potential sexual barriers between populations and could facilitate ecological modeling of sexual reproduction and speciation. Targeted sampling of insertion sequences in other eukaryotic lineages, and viruses, cultured and wild, should lead to further discovery of novel insertion elements and shed light on the origins of those observed here.
Acknowledgments
We thank H Moreau and E Derelle for the BBan7 PRP8 sequence, M Butler for Spu-PRP8 and B Marin for providing the Mamiellophyceae 18S, ITS alignment. We also thank Q Eastman, MP Simmons, R Gausling and three anonymous reviewers for constructive criticism and editing. This research was supported by the Lucille and David Packard Foundation and grants from the GBMF, NSF (0843119) and DOE to AZW.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Bokor AA, Kohn LM, Poulter RT, van Kan JA. PRP8 inteins in species of the genus Botrytis and other ascomycetes. Fungal Genet Biol. 2012;49:250–261. doi: 10.1016/j.fgb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Burt A, Koufopanou V. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr Opin Genet Dev. 2004;14:609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Butler MI, Gray J, Goodwin TJ, Poulter RT. The distribution and evolutionary history of the PRP8 intein. BMC Evol Biol. 2006;6:42. doi: 10.1186/1471-2148-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley AI, Asuncion BF, Steward GF. Detection of inteins among diverse DNA polymerase genes of uncultivated members of the Phycodnaviridae. ISME J. 2009;3:409–418. doi: 10.1038/ismej.2008.120. [DOI] [PubMed] [Google Scholar]
- Cuvelier ML, Allen AE, Monier A, McCrow JP, Messié M, Tringe SG, et al. Targeted metagenomics and ecology of globally important uncultured eukaryotic phytoplankton. Proc Natl Acad Sci USA. 2010;107:14679–14684. doi: 10.1073/pnas.1001665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir-Hilton E, Sudek S, Cuvelier ML, Gentemann C, Zehr JP, Worden AZ. Global distribution patterns of distinct clades of the photosynthetic picoeukaryote Ostreococcus. ISME J. 2011;5:1095–1107. doi: 10.1038/ismej.2010.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic M, Mushegian A. PRP8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse tranascriptase. RNA. 2012;17:799–808. doi: 10.1261/rna.2396011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J.2005PHYLIP (Phylogeny Inference Package) version 3.6.3.6 ednsDistributed by the authorDepartment of Genome Sciences, University of Washington: Seattle, WA, USA [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Senejani AG, Zhaxybayeva O, Olendzenski L, Hilario E. Inteins: structure, function, and evolution. Annu Rev Microbiol. 2002;56:263–287. doi: 10.1146/annurev.micro.56.012302.160741. [DOI] [PubMed] [Google Scholar]
- Goodwin TJ, Butler MI, Poulter RT. Multiple, non-allelic, intein-coding sequences in eukaryotic RNA polymerase genes. BMC Biol. 2006;4:38. doi: 10.1186/1741-7007-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hurst GD, Werren JH. The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet. 2001;2:597–606. doi: 10.1038/35084545. [DOI] [PubMed] [Google Scholar]
- Irimia M, Roy SW. Evolutionary convergence on highly-conserved 3' intron structures in intron-poor eukaryotes and insights into the ancestral eukaryotic genome. PLoS Genet. 2008;4:e1000148. doi: 10.1371/journal.pgen.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Tucker AE, Sung W, Thomas WK, Lynch M. Extensive, recent intron gains in Daphnia populations. Science. 2009;326:1260–1262. doi: 10.1126/science.1179302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Intron evolution as a population-genetic process. Proc Natl Acad Sci USA. 2002;99:6118–6123. doi: 10.1073/pnas.092595699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin B, Melkonian M. Molecular phylogeny and classification of the Mamiellophyceae class. nov. (Chlorophyta) based on sequence comparisons of the nuclear- and plastid-encoded rRNA operons. Protist. 2010;161:304–336. doi: 10.1016/j.protis.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Moliner C, Fournier PE, Raoult D. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol Rev. 2010;34:281–294. doi: 10.1111/j.1574-6976.2010.00209.x. [DOI] [PubMed] [Google Scholar]
- Monier A, Welsh RM, Gentemann C, Weinstock G, Sodergren E, Armbrust EV, et al. Phosphate transporters in marine phytoplankton and their viruses: cross-domain commonalities in viral-host gene exchanges. Environ Microbiol. 2012;14:162–176. doi: 10.1111/j.1462-2920.2011.02576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki K, Shirai Y, Tomaru Y, Nishida K, Pietrokovski S. Algal viruses with distinct intraspecies host specificities include identical intein elements. Appl Environ Microbiol. 2005;71:3599–3607. doi: 10.1128/AEM.71.7.3599-3607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols SA, Roberts BW, Richter DJ, Fairclough SR, King N. Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/beta-catenin complex. Proc Natl Acad Sci USA. 2012;109:13046–13051. doi: 10.1073/pnas.1120685109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, La Scola B, Audic S, Renesto P, Blanc G, Robert C, et al. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2006;2:e76. doi: 10.1371/journal.pgen.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Raoult D, Claverie JM. A new example of viral intein in Mimivirus. Virol J. 2005;2:8. doi: 10.1186/1743-422X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owczarzak A, Stibbs HH, Bayne CJ. The destruction of Schistosoma mansoni mother sporocysts in vitro by amoebae isolated from Biomphalaria glabrata: an ultrastructural study. J Invertebr Pathol. 1980;35:26–33. doi: 10.1016/0022-2011(80)90079-8. [DOI] [PubMed] [Google Scholar]
- Perler FB. InBase: the intein database. Nucleic Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel W, Chan L, Dominus M, Ruan J, Vos R, V T. TreeBASE v. 2: A Database of Phylogenetic Knowledge. e-BioSphere; 2009: London, UK; 2009. [Google Scholar]
- Pietrokovski S. Intein spread and extinction in evolution. Trends Genet. 2001;17:465–472. doi: 10.1016/s0168-9525(01)02365-4. [DOI] [PubMed] [Google Scholar]
- Poulter RT, Goodwin TJ, Butler MI. The nuclear-encoded inteins of fungi. Fungal Genet Biol. 2007;44:153–179. doi: 10.1016/j.fgb.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Carmel L, Csuros M, Koonin EV. Origin and evolution of spliceosomal introns. Biol Direct. 2012;7:11. doi: 10.1186/1745-6150-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol. 2008;25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. J Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skippington E, Ragan MA. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol Rev. 2011;35:707–735. doi: 10.1111/j.1574-6976.2010.00261.x. [DOI] [PubMed] [Google Scholar]
- Swithers KS, Senejani AG, Fournier GP, Gogarten JP. Conservation of intron and intein insertion sites: implications for life histories of parasitic genetic elements. BMC Evol Biol. 2009;9:303. doi: 10.1186/1471-2148-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaulot D, Lepere C, Toulza E, De la Iglesia R, Poulain J, Gaboyer F, et al. Metagenomes of the picoalga bathycoccus from the Chile coastal upwelling. PLoS One. 2012;7:e39648. doi: 10.1371/journal.pone.0039648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ. Picoeukaryote diversity in coastal waters of the Pacific Ocean. Aquat Microb Ecol. 2006;43:165–175. [Google Scholar]
- Worden AZ, Lee JH, Mock T, Rouze P, Simmons MP, Aerts AL, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- Yahara K, Fukuyo M, Sasaki A, Kobayashi I. Evolutionary maintenance of selfish homing endonuclease genes in the absence of horizontal transfer. Proc Natl Acad Sci USA. 2009;106:18861–18866. doi: 10.1073/pnas.0908404106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.