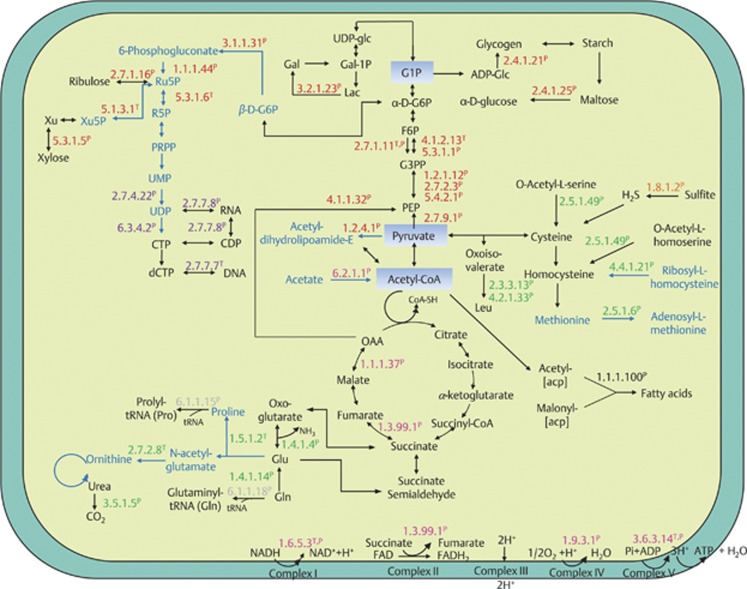
Figure 4.
Metabolic map of D. colotermitum strain TAV2 constructed with selected differentially expressed genes and proteins. Proteins (P) and/or their mRNA transcripts (T) are classified into six major metabolic classes: carbohydrate metabolism (red), energy generation (pink), amino-acid biosynthesis (green), nucleotide metabolism (purple), fatty acid synthesis (black) and translation (gray). The enzyme sulfite reductase is shown in orange. Text marked in black represents upregulated pathways under 2% O2, whereas text marked in blue represents downregulated pathways. acp, acyl carrier protein; ADP, adenosine diphosphate; ADP-glc, adenosine diphosphoglucose; CTP, cytidine triphosphate; CDP, cytidine diphosphate; dCTP, deoxycytidine triphosphate; Gal-1P, galactose-1-phosphate; Gal, galactose; G1P, glucose-1-phosphate; G6P, glucose-6-phosphate; G3PP, glyceraldehyde-3-phosphate; F6P, fructose-6-phosphate; FAD, flavin adenine dinucleotide (oxidized form); FADH, flavin adenine dincleotide (reduced form); Lac, lactose; Leu, leucine; Glu, glutamate; Gln, glutamine; NAD, nicotinamide adenine dinucleotide (oxidized form); NADH, nicotinamide adenine dinucleotide (reduced form); OAA, oxaloacetate; PEP, phosphoenolpyruvate; PRPP, phosphoribosyl pyrophosphate; Ru5P, ribulose 5-phosphate; R5P, ribose 5-phosphate; UDP-glc, uridine diphosphate glucose; UMP, uridine monophosphate; UDP, uridine diphosphate.