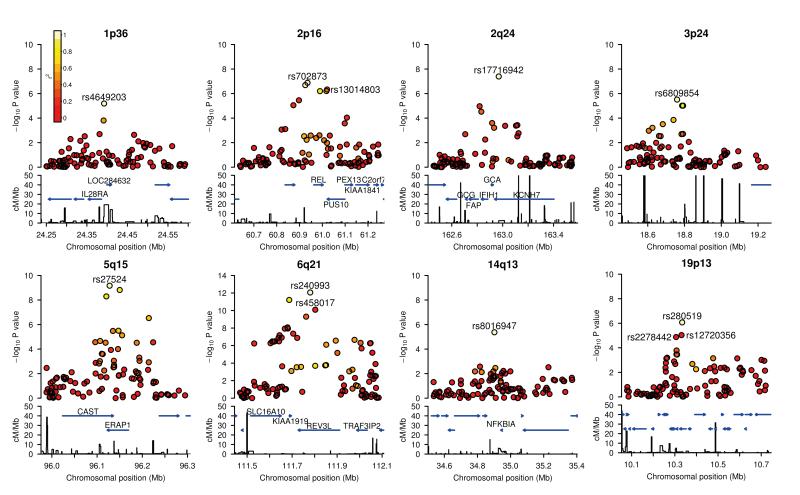
Figure 2.
Regional association plots.
The -log10 (P) values for SNPs at eight new loci are shown on the upper part of each plot. SNPs are coloured based on their r2 with the labeled hit SNP which has the smallest P value in the region. r2 is calculated from the 58C control data. Where more than one SNP is labeled, there is evidence for multiple signals at the locus (see text). The bottom section of each plot shows the fine scale recombination rates estimated from HapMap individuals and genes are marked by horizontal blue lines. Genes within the recombination region of the hit SNP are labeled except for 19p13, where the genes are: GLP-1, FDX1L, RAVER1, ICAM3, TKY2, CDC37, PDE4A, KEAP1, S1PR5.