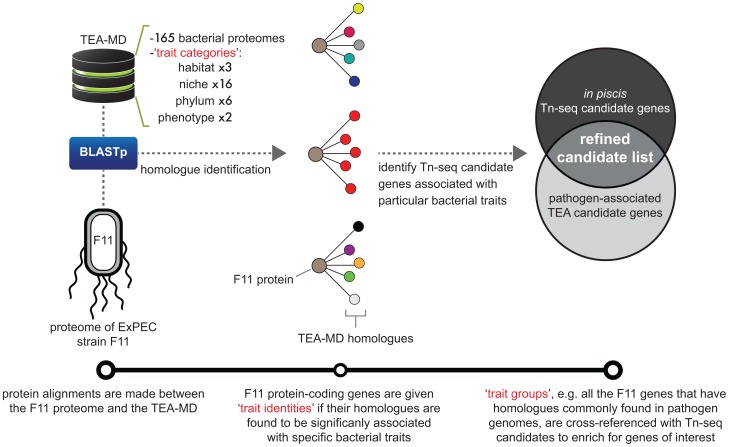
Figure 5. Diagrammatic workflow of ‘Trait Enrichment Analysis’ (TEA).
This visualization depicts the TEA method and describes the relationships between various data types. Bottom track: linear representation of steps followed in the execution of TEA. Protein alignments are made between F11 proteins and the TEA-MD to identify homologues. Microbes within the TEA-MD are annotated with four separate traits contained within the trait categories: habitat (1 of 3), niche (1 of 16), phylum (1 of 6) and phenotype (1 of 2). The composition of traits associated with homologue sets for each F11 protein is assessed for enriched traits in order to assign trait identities. Trait groups, which are F11 genes that share a particular trait identity, are used to organize Tn-seq-derived candidate genes. TEA-MD = TEA-metaproteome database; BLASTp = Basic Local Alignment Search Tool used to align protein sequences.