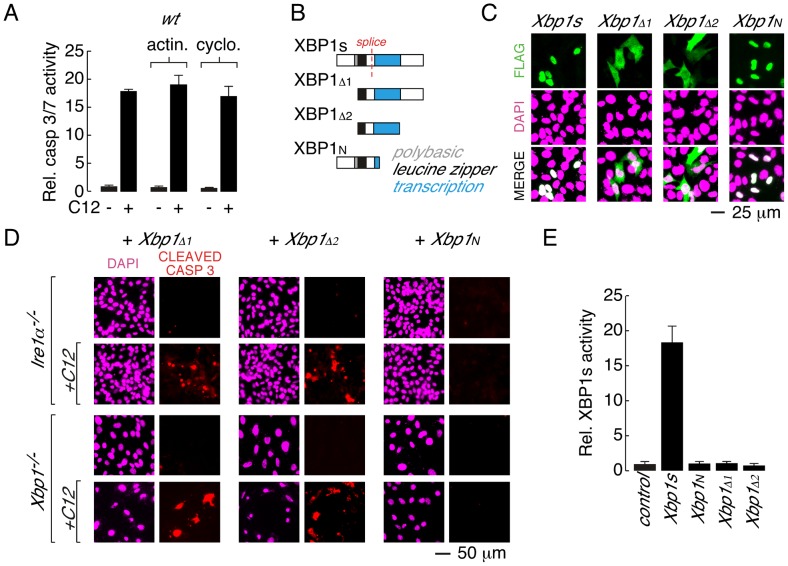
Figure 4. Leucine zipper and transcriptional activation domains of XBP1s are sufficient for C12-mediated caspase activation.
A. Acute inhibition of RNA or protein synthesis with actinomycin (actin.) or cycloheximide (cyclo.) does not inhibit C12-induced caspase activation in wt MEFs. B. Domain structure of XBP1s and XBP1s truncations. XBP1s contains a polybasic domain (grey), leucine zipper domain (black) and transcriptional activation domain (blue). The location of the splice site that converts XBP1u pre-mRNA to XBP1s mRNA is also shown (red). The FLAG-tag on all constructs is located at the amino-terminus. C. Analysis of cellular localization of XBP1s and XBP1s truncations by immunocytochemistry. Panels are shown for anti-FLAG immunostaining (top), DAPI staining (middle), and merged images (bottom). D. Expression of XBP1s truncations that contain the leucine zipper and transcriptional activation domains (XBP1Δ1 and XBP1Δ2) is sufficient to restore C12-mediated caspase activation in Ire1α −/− (top panels) and Xbp1−/− (bottom panels) MEFs. Expression of XBP1N failed to restore C12-mediated caspase activation. Images are shown for DAPI staining (magenta) and caspase immunostaining (red) in the absence and presence of C12. E. XBP1s truncations that restore C12-mediated caspase activation (XBP1Δ1 and XBP1Δ2) are transcriptionally inactive. Normalized XBP1s transcriptional activity was measured in wt MEFs using an XBP1s-responsive luciferase-based reporter and co-expression of control plasmid, XBP1s, or XBP1 truncations. Individual scale bars are shown for panels C and D.