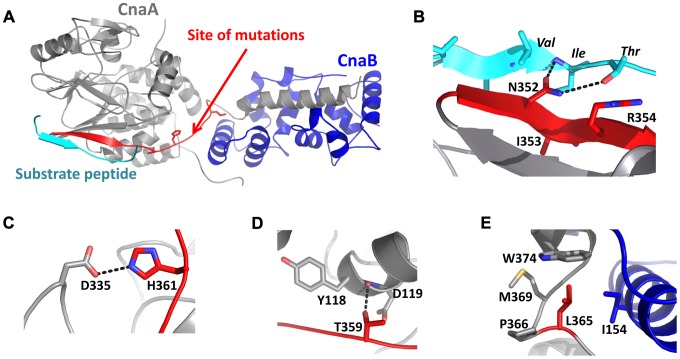
Figure 12. Molecular modeling of A.
fumigatus CnaA substrate recognition motif. (A) Model of the A. fumigatus CnaA (gray) and CnaB (blue) subunits, indicating relative positions of substrate peptide (cyan) and sites on the protein where mutations are found (red). The Phyre server was used to prepare homology models using the X-ray structure of the human calcineurin heterodimer bound to a substrate peptide (PDB ID 2P6B). Most mutations are predicted to destabilize portions of the protein required for heterodimer formation or substrate recognition. (B) Interactions of CnaA subunit with substrate peptide of sequence PVIVIT. N352A mutation would disrupt several hydrogen bonds that stabilize this interaction. (C) H361 forms interactions (including a likely hydrogen bond with D335) that are important for maintaining the structure of CnaA near the substrate binding region. H361L mutation would abolish these interactions and place a hydrophobic residue in a fairly solvent-exposed portion of the protein. (D) As with H361, T359 is important for structural stability near the substrate binding region of CnaA. T359P mutation would disrupt these important interactions. (E) L365 lies in a hydrophobic pocket at the interface between CnaA and CnaB subunits; L365S mutation would be unfavorable for heterodimer formation.