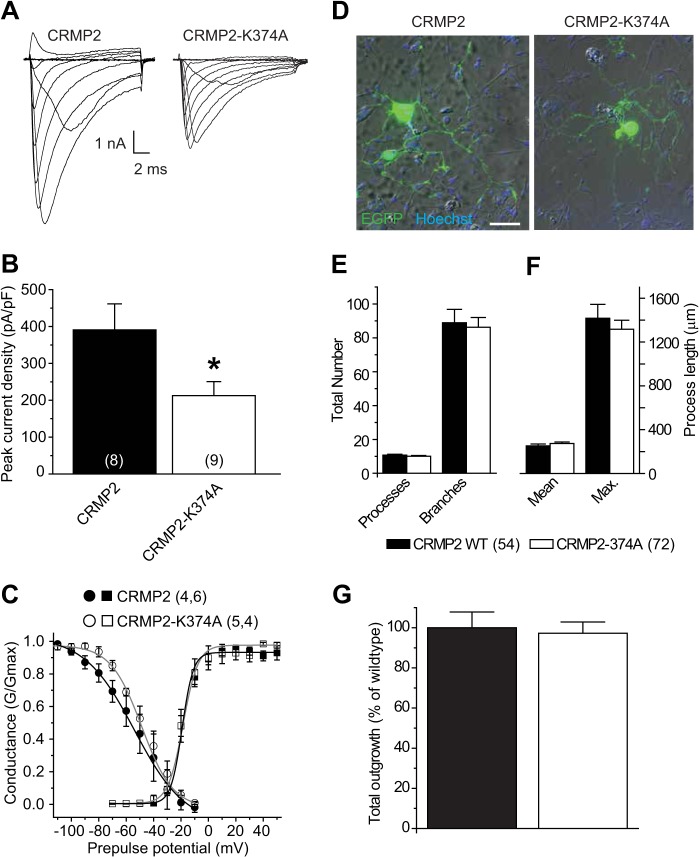
FIGURE 11.
CRMP2-K374A mutant reduces sodium current density in DRG neurons but does not impair CRMP2-mediated neurite outgrowth. A, representative sodium currents recorded in dissociated rat DRG neurons 48 h after transfection with EGFP and CRMP2 (left) or EGFP and CRMP2-K374A (right). B, summary bar graph showing peak sodium current in each condition. Numbers in parentheses represent numbers of cells tested. There was a significant reduction in peak current density in DRG neurons expressing CRMP2-K374A compared with those expressing wild type CRMP2 (p < 0.05, Student's t test). C, representative Boltzmann fits for activation and steady-state inactivation for DRG neurons transfected with the indicated constructs are shown. The calculated values for V½ and k of activation were not significantly different between the conditions as follows: −19.2 ± 0.8 mV and 3.9 ± 0.4 mV/e-fold (n = 6) for EGFP plus CRMP2-transfected cells versus −18.5 ± 1.3 mV and 5.5 ± 0.7 mV/e-slope (n = 4) for EGFP plus CRMP2-K374A-transfected cells (p > 0.05 for both parameters, Student's t test). CRMP2-K374A produced a significant ∼9-mV depolarizing shift in V½ of steady-state inactivation: −59.9 ± 2.2 mV for EGFP plus CRMP2-transfected cells versus −50.7 ± 1.8 mV for EGFP and CRMP2-K374A expressing cells (n = 4 for both conditions, p < 0.05, Student's t test). Values for k of steady-state inactivation were not different between conditions: 13.2 ± 1.3 mV/e-fold for EGFP plus CRMP2-transfected cells versus 10.6 ± 2.0 for EGFP plus CRMP2-K374A-transfected cells (n = 4 for both conditions, p > 0.05, Student's t test). D, example image showing the overlay of EGFP fluorescence (green) and nuclear staining with Hoechst (blue) together with the brightfield Hoffman image of transfected DRG neurons. Scale bar equals 50 μm and applies to both panels. Note the similar degree of neurite complexity between the wild type and mutant CRMP2 conditions. E, number of processes and branches; F, mean and maximum (max.) process length, and G, normalized total outgrowth for cells co-expressing EGFP and CRMP2 (n = 54 from four wells) or co-expressing EGFP and CRMP2-K374A (n = 72 from four wells). There were no statistical differences for any of the parameters between the two conditions (p > 0.05, Student's t test).