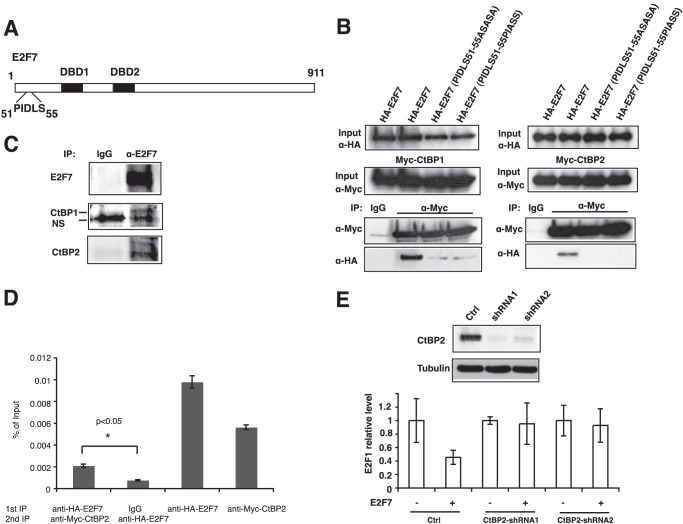
FIGURE 4.
E2F7 mediates transcription repression through CtBPs. A, illustration of potential CtBP-binding motif (PIDLS) located at the N terminus of E2F7. B, E2F7 interacts with CtBP1 and CtBP2. 293 cells were transfected with overexpression plasmids encoding Myc-tagged CtBPs and HA-E2F7 as indicated. Cell lysate was immunoprecipitated (IP) with anti-Myc antibody followed by immunoblotting with anti-HA or anti-Myc antibodies as indicated. Immunoprecipitation with normal mouse IgG was used as negative control. C, endogenous E2F7 interacts with CtBPs. Lysate from HeLa cells was immunoprecipitated (IP) with anti-E2F7 antibody followed by immunoblotting with anti-CtBP1 or anti-CtBP2 antibody as indicated. Note that the lower band of CtBP1 blot is nonspecific (NS). D, ChIP-reChIP assays. 293 cells were co-transfected with HA-E2F7 and Myc-CtBP2. Cells were cross-linked after 24 h, lysed, sonicated, precleared, and immunoprecipitated with the indicated antibodies overnight followed by incubation with protein A/G beads for 2 h. After stringent washes, chromatin-bound proteins were eluted with 1 mm dithiothreitol followed by a second immunoprecipitation with the indicated antibodies. Chromatin was eluted from beads, uncross-linked, purified, and subjected to real time PCR for E2F1 promoter. Two-tailed t test was performed for statistical analysis. The asterisk indicates that the difference is significant (p < 0.05). E, CtBP2 is required for E2F7-mediated repression. 293T cells (5 × 105 cells) were infected with lentivirus harboring short hairpin RNAs (shRNA) targeting CtBP2 (two different hairpin sequences per gene were chosen). Cells were selected with 2 μg/ml puromycin for 48 h, transfected with 500 ng of HA-E2F7 for 14 h, and harvested for assays. Western blot was conducted to confirm the depletion of CtBP2 proteins (upper panel). RNA was isolated and reverse-transcribed, and the E2F1 level was measured by real time PCR. Cell line integrated with a nonsilencing shRNA was used as a control (Ctrl). All values across samples were normalized with glyceraldehyde phosphate dehydrogenase level.