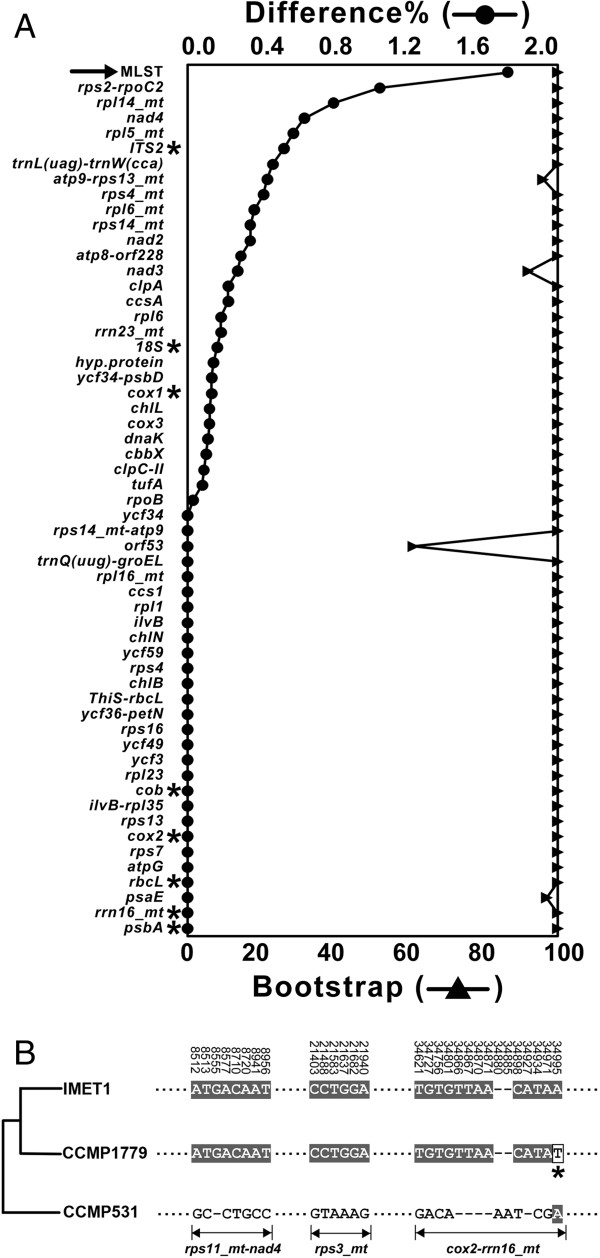
Figure 5.
Multiple locus sequence tag for high-resolution phylotyping of three closely related N. oceanica strains. (A) Comparison of the sensitivity and specificity among candidate regions/markers for intra-species discrimination. The regions/markers derived from genic and intergenic sequences of pt and mt of the N.oceanica strains (IMET1 and CCMP531) were listed on the X axe. The % nucleotide difference of each region/markers was calculated as the index for sensitivity. The bootstrap values of the IMET1-CCMP531 branches in the sequence-specific phylogenetic trees (maximum parsimony; MP) were shown as the index for specificity. *: presently used markers. Arrow: the three regions (rps11_mt-nad4, rps3_mt and cox2-rrn16_mt) for Multiple-Locus Sequence Typing (MLST). (B) Phylogeny of IMET1, CCMP531 and CCMP1779 as reconstructed from three MLST loci. Grey background: identical loci in two strains. Blank box and *: A base that is different between CCMP1779 and IMET1.