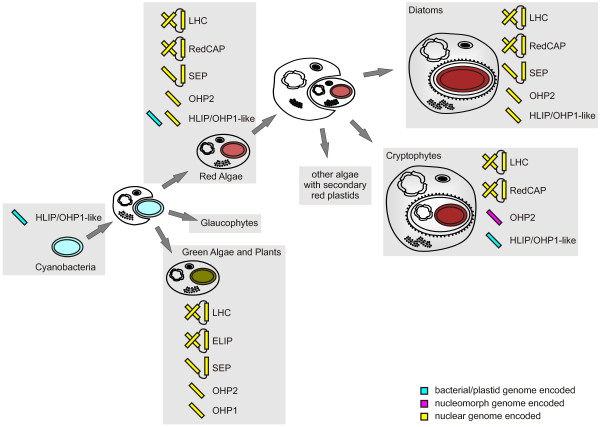
Figure 2.
Proposed evolutionary history of RedCAP, LHC and LHC-like genes. RedCAPs evolved in red algae after primary endocytobiosis and their genes can be found in the nuclear genome of almost all red algae investigated so far. During secondary endocytobiosis, when a red alga was taken up by another eukaryotic host, the RedCAP gene was transferred to the nucleus of the host cell and was lost from the nuclear genome of the former endosymbiont. Similarly, different nuclear encoded LHC variants evolved in organisms with primary plastids and were transferred to the nucleus of the secondary host during the evolution of secondary plastids. Interestingly, in diatoms, HLIP/OHP1-like genes (plastid encoded in red algae) as well as OHP2 genes (nucleus encoded in red algae) have been transferred to the nucleus of the secondary host cell. This is in contrast to the situation in the cryptophyte Guillardia theta, in which HLIP/OHP1-like is plastid encoded, and OHP2 is nucleomorph encoded. Table S2 (see Additional file 7) contains detailed genome/gene information.