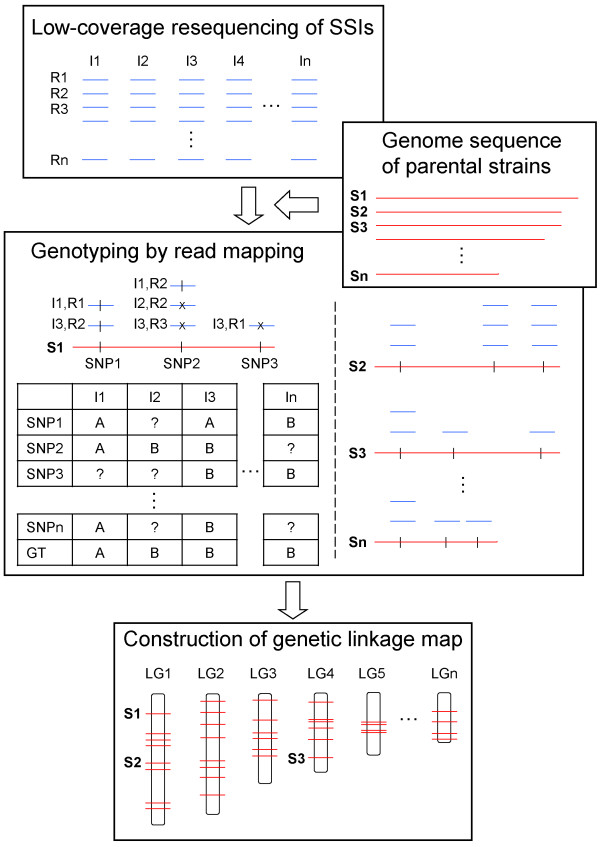
Figure 2.
Flowchart of the proposed genotyping approach. Low-coverage (~0.5 to 1-fold) resequencing of a selected mapping population, here single-spore isolates (SSIs) I1 to In, is first performed on a next-generation sequencing (NGS) platform such as Roche 454, generating a library of short shotgun reads R1 to Rn for each isolate. Genotyping of the SSIs is then carried out by mapping the reads of each SSI to the draft parental reference genomes, represented in scaffolds S1 to Sn here. On each scaffold, identical mapped reads are assigned the same genotype of the parental strain being mapped whereas reads with high-quality single-nucleotide polymorphisms (SNPs) that are present in the other parent are assigned the opposite genotype. For each SSI, the final genotype (GT) of each scaffold, serving as a genetic marker, is called by a simple majority vote. All the scaffolds are genotyped in this manner. A genetic linkage map, on which linkage information of the markers on a set of linkage groups (LGs) is displayed, is then built based on the genotype segregation ratio of every marker examined.