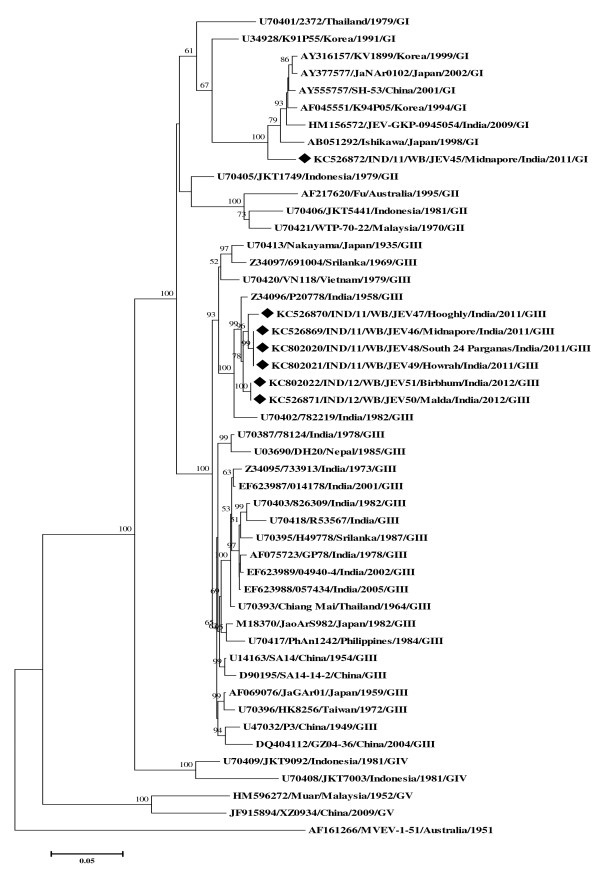
Figure 1.
Phylogenetic relationship among JEV isolates from WB, India. The Neighbor-Joining (NJ) Phylogenetic tree, tested with Kimura 2-parameter model was generated by MEGA5, using the complete E gene nucleotide sequences of 4 JEV isolates from hospitalized AES case-patients in West Bengal during 2011–12, with reference to other 40 wild type JEV strains from worldwide. The strain MVEV-1-51 was used as an out group for generating the rooted tree. The ≥50% bootstrap support values (1000 pseudo replicates) were shown in corresponding nodes. Horizontal branch lengths are proportional to genetic distance and vertical branch lengths have no significance. Each taxon is named systematically by mentioning the accession number, strain/isolate name, country of origin, year of isolation and genotype. The isolates’ sequences used in this study were indicated by ‘♦’ mark. Scale bar indicates nucleotide substitutions per site.