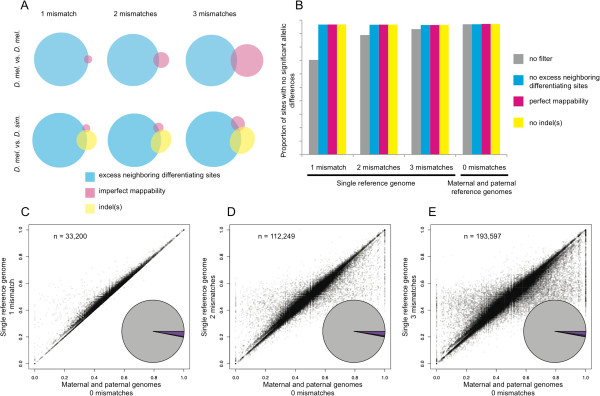
Figure 5.
Real reads aligned to a single reference genome produce reliable measures of allelic abundance after excluding problematic differentiating sites. (A) The relative proportions of sites with an excess of neighboring differentiating sites (cyan), imperfect mappability (magenta), an indel(s) nearby (yellow), or more than one of these properties are shown for the simulated 36-base intra- (mel-mel) and interspecific (mel-sim) datasets allowing one (1 mm), two (2 mm), or 3 (3 mm) mismatches during alignment to a single reference genome. (B) The proportion of differentiating sites with no statistically significant difference in relative allelic expression is shown for the real reads from F1 hybrids between D. melanogaster and D. simulans after aligning to either a single reference genome with one, two, or three mismatches allowed or to both the maternal and paternal genomes with zero mismatches allowed before excluding any sites (grey) and after sequentially excluding differentiating sites with an excess of neighboring differentiating sties (cyan), imperfect mappability (magenta), or an indel(s) nearby (yellow). (C-E) For each differentiating site retained after filtering based on neighboring differentiating sites, mappability, and indels, the proportion of reads assigned to the reference allele is plotted after aligning reads to a single reference genome (y-axis) or to separate allele-specific genomes (x-axis), allowing one (C), two (D), or three (E) mismatches. The pie chart insets reflect the total number of differentiating sites that showed either no statistically significant difference in relative allelic abundance using either alignment strategy (grey), a statistically significant difference when reads were aligned to either a single reference genome (blue) or both the maternal and paternal genomes (red), or a significant difference with both alignment methods (purple). Binomial exact tests and a false discovery rate of 0.05 were used to assess statistical significance in all cases.