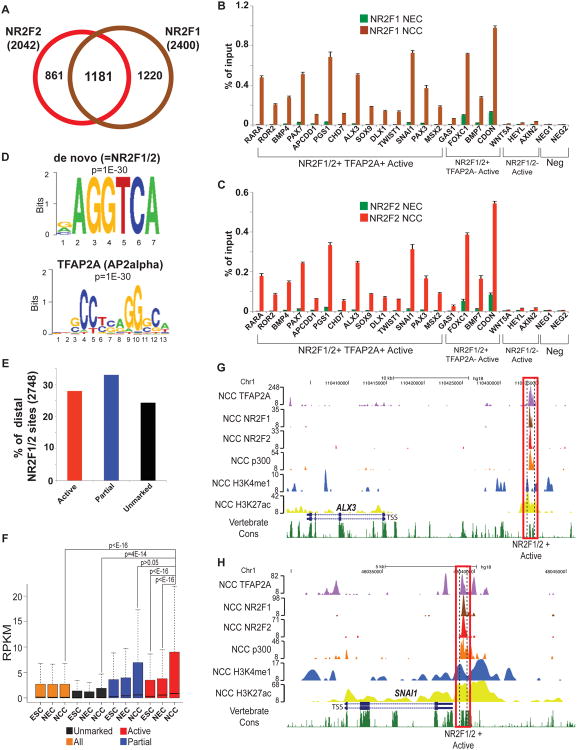
Figure 5. Genome-wide analysis of NR2F1/2 occupancy in hNCC (see also Figure S6).
(A) Overlap between NR2F1 (brown) and NR2F2 (red) binding sites in hNCC. (B-C) Chromatin isolated from hNCC and neurectodermal spheres (hNEC), just prior to attachment and emigration of hNCC was used for NR2F1 (B) and NR2F2 (C) ChIP-qPCRs. Error bars represent standard deviation from three technical replicates. (D) Top overrepresented motifs enriched at distal NR2F1/2-bound regions based on either de novo motif analysis (top, which matches NR2F1/2 motif) or matches to known transcription factors (bottom). (E) Percentage of distal NR2F1/2 sites that overlapped active hNCC enhancers (red), regions with a partial active enhancer signature (blue) or that occurred within unmarked chromatin (black). (F) Expression levels, measured as RPKMs, were calculated for all human ENSEMBL genes and for those closest (within 100 Kb) to distal NR2F1/2 bound regions overlapping active hNCC enhancers (red), or displaying a partial active enhancer signature (blue) or occurring within unmarked chromatin (black). Expression levels presented as boxplots. P-values calculated using Wilcoxon tests. (G-H) ChIP-seq enrichment profiles in hNCC for two representative NR2F1/2-bound loci (proximal to ALX3 (G) and SNAI1 (H)) overlapping active hNCC enhancers.