Abstract
Notch receptors are single-pass transmembrane proteins that regulate development and tissue homeostasis in all metazoan organisms. Prior to ligand-induced signaling, Notch receptors adopt a proteolytic-resistant conformation maintained by a critical interdomain interface within a negative regulatory region (NRR), which sits immediately external to the plasma membrane. Signaling is initiated when ligand binding induces exposure of the proteolytic cleavage site, termed S2, within the NRR. Here, we use hydrogen exchange in conjunction with mass spectrometry (HX-MS) to study the dynamics of the human Notch3 NRR in four distinct biochemical states: in its unmodified quiescent form, in a proteolytically “on” state induced by EDTA, and in complex with either agonist or inhibitory antibodies. Induction of the “on” state by either EDTA or the agonist monoclonal antibody leads to accelerated deuteration in the region of the S2 cleavage site, reflecting an increase in S2 dynamics. In contrast, complexation of the Notch3 NRR with an inhibitory antibody retards deteuration not only across its discontinuous binding epitope, but also around the S2 site, stabilizing the NRR in its “off” state. Together with previous work investigating the dynamics of the Notch1 NRR, these studies show that key features of autoinhibition and activation are shared among different Notch receptors, and provide additional insights into mechanisms of Notch activation and inhibition by modulatory antibodies.
Introduction
Notch signaling is a highly conserved pathway that influences cell fate decisions during embryonic development and tissue homeostasis of metazoan organisms. Precise regulation of Notch pathway activity is crucial, as increases or deficiencies of signaling are associated with developmental disorders 1,2, neurological diseases 3, and a wide range of human cancers 4,5.
Notch receptors constitute a family of single-pass transmembrane proteins that signal via contact with their ligands on neighboring cells. Mammals have four Notch receptors (Notch1 to Notch4), which share a similar modular organization (Figure 1). The large extracellular portion of the receptor encompasses a series of epidermal growth factor (EGF)-like repeats that are responsible for ligand binding 6, followed by a negative regulatory region (NRR) that maintains the receptor in a quiescent protease-resistant state prior to ligand stimulation 7-10. The NRR consists of three highly conserved LIN-12/Notch repeats (LNRs) and a heterodimerization domain (HD). The HD domain contains two proteolytic cleavage sites termed S1 and S2. The S1 site is typically processed by furin-like protease during maturation 11,12, and the S2 site is cleaved by ADAM-type metalloproteases in response to ligand stimulation 13,14.
Figure 1.
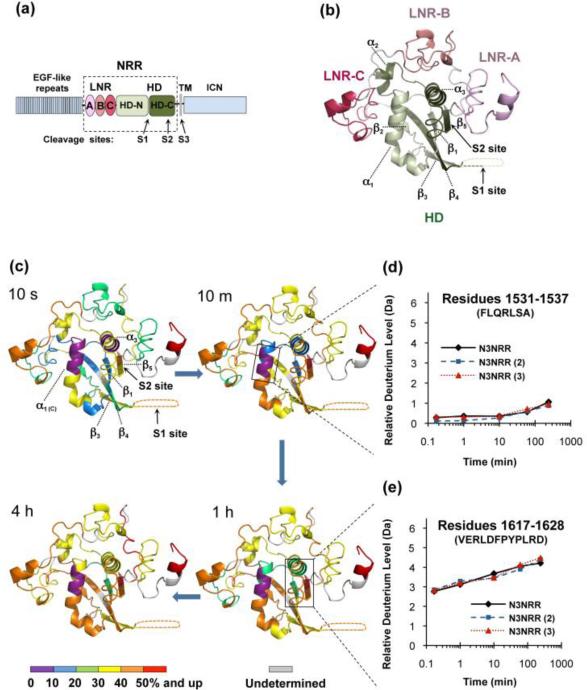
HX-MS of Notch3 NRR in its autoinhibited state. (a) Domain organization of human Notch3. The extracellular portion is comprised of 34 EGF-like repeats and the negative regulatory region (NRR, boxed). The NRR consists of three LNR repeats (LNR-A, LNR-B and LNR-C, colored in different shades of pink) and an HD domain (N-terminus and C-terminus, colored in different shades of green). The Notch transmembrane domain (TM), the intracellular region (ICN), and the three key proteolytic cleavage sites (S1, S2 and S3) are also indicated. (b) Homology model of the human Notch3 NRR, derived from the crystal structure of the Notch 2 NRR (pdb: 2OO4) using Phyre. The various domains are colored as in (a). Important secondary structural elements and key cleavage sites (S1 and S2) are also noted. (c) Time course showing the HX-MS behavior of the Notch3 NRR in its “off” state. The ribbon diagram is colored according to the relative extent of deuteration of peptides mapped to the indicated regions of the Notch3 NRR. The color code is indicated at the bottom of the figure. Sequences for which HX-MS data could not be acquired are colored gray. (d) Deuterium incorporation plot of the C-terminal region of helix one (α1), which is among the most highly protected structural elements of the Notch3 NRR. (e) Deuterium incorporation plot of a peptide that includes the S2 cleavage site. Data for both graphs are shown for three independent experiments to emphasize the reproducibility of the measurements. The full dataset for all peptides is shown in Figure S4A.
S2 cleavage is the crucial committed step in both normal and pathophysiologic activation of Notch receptors. The S2-processed receptor is further processed by gamma secretase within the Notch transmembrane region at S3 (and additional sites) 15,16. After gamma secretase cleavage, the intracellular part of Notch (ICN) is released from the membrane, and translocates into the nucleus where it forms a transcriptional activation complex involved in regulation of Notch-responsive genes 17,18.
The importance of Notch signaling in human biology is highlighted by well-established associations between Notch receptor mutations and several human diseases. In human T-cell acute lymphoblastic leukemia (T-ALL), more than 50% of patients have mutations in Notch1 that lead to aberrant increases in Notch activity 19. On the other hand, loss-of-function mutations in the Notch2 receptor (and of the canonical ligand Jagged1) are associated with Alagille syndrome 1,2. Inherited mutations of Notch3 lead to a hereditary stroke syndrome known as CADASIL 3 (Congenital Autosomal Dominant Arteriopathy with Strokes, Infarcts, and Leukencephalopathy), and somatic amplification of the Notch3 locus is observed in approximately 20% of serous ovarian adenocarcinomas 20,21. Thus, a detailed investigation of the mechanism of receptor autoregulation is warranted for this important family of signaling proteins.
We previously 22 investigated the dynamics of the Notch1 NRR in autoinhibited and activated states using hydrogen-exchange mass-spectrometry (HX-MS) 23. Those studies also analyzed the inhibitory effects of an allosteric antibody that binds directly to the NRR, stabilizing it in the “off” state 24. That work helped shed light on the signaling mechanisms of human Notch1, and laid the foundation for subsequent HX-MS studies of other Notch receptors.
In addition to the inhibitory antibodies directed against the Notch1 NRR, there are also modulatory antibodies that bind to the NRR regions of Notch2 and Notch3 25,26. In particular, the antibodies directed against the Notch3 NRR provide a unique opportunity to investigate the effects of antibody modulation of Notch signaling activity, because both inhibitory and activating antibodies have been developed for potential therapeutic applications in cancer and other diseases, and their epitopes previously characterized using biochemical methods 25.
Here, we utilize HX-MS to investigate the effects of both chemical and antibody-induced perturbations of the Notch3 NRR that lead to Notch3 receptor activation and inhibition. Comparison of the exchange properties of the Notch1 and Notch3 NRRs indicates that the main biochemical features of autoinhibiton, including retarded deuteration of the S2 site, are preserved in the two “off” states. Induction of the “on” state by either EDTA or an agonist monoclonal antibody leads to accelerated deuteration in the region of the S2 cleavage site, reflecting an increase in S2 accessibility. In contrast, complexation of the Notch3 NRR with an inhibitory antibody retards the deteuration around the S2 site, stabilizing the NRR in its “off” state.
Results
Dynamics of the NRR from Human Notch3
In this study, we sought to gain a molecular understanding of the dynamics and conformational transitions of the human Notch3 NRR. We first confirmed that the Notch3 NRR is indeed autoinhibited in isolation. Then, we used HX-MS to investigate the dynamics of the Notch3 NRR in its autoinhibited or “off” state in order to compare it with the Notch1 NRR. To explore the dynamics of the Notch3 NRR by HX-MS, the recombinant NRR was first incubated in a deuterated buffer for various periods of time, ranging from 10 seconds to 4 hours. The exchange reactions were quenched by adjusting the pH to 2.6 at 0 °C. The quenched reaction mixture was digested with pepsin, and the digested peptides were separated by UPLC (see Figure S2 for peptide map). The deuterium incorporation level of each peptidic fragment at each time point was determined by mass spectrometry, and the HX-MS data for the various peptides were then mapped onto a structural model of the Notch3 NRR (Figure 1b), which was built based on the high-resolution crystal structure of the Notch2 NRR in its autoinhibited conformation using Phyre 27,28.
Overall, the HX-MS results of the Notch3 NRR in its autoinhibited state support the architectural features of the molecular model, with the three LNR repeats wrapping around the compact HD domain to protect the S2 site. Our data show that the periphery of the NRR, including the three LNR repeats, is more rapidly deuterated than the HD hydrophobic core (Figure 1c and Figure S4A). The interior of the HD domain, especially the C-terminal part of helix one (α1), is more resistant to deuteration (Figure 1c, d). The peptide in this region exchanges fewer than 20% of its backbone amide protons even after four hours of deuterium labeling. These Notch3 results are consistent with previous studies of Notch1 (Figure S3), which showed helix α1 to be among the most protected regions of the NRR 22. Notably, many of the leukemia-associated mutations in the Notch1 NRR are found in this α1 helix 10. This finding therefore emphasizes the important role of helix α1 in maintaining the structural integrity of other Notch NRRs as well.
Dynamics around the S2 site
The metalloprotease cleavage site is located within strand β5 of the HD domain, which immediately precedes the transmembrane region (Figure 1e, 2). The major peptide spanning this region (residues 1617-1628) also includes a linker that connects helix α3 to strand β5, and that is likely to be flexible given the flexibility of this region in the Notch1 NRR and the structural divergence of this region between Notch1 and Notch2 10. This peptide shows substantial deuteration even at early time points, with additional deuterium incorporation as a function of time. Because the exchange dynamics of this peptide report on both strand β5 and its adjacent linker, this pattern is most consistent with an interpretation in which the linker exchanges rapidly, and strand β5 exhibits protection by comparison to the exposed linker region. This interpretation is buttressed by analysis of the exchange behavior of the other secondary structural elements surrounding the S2 site, strand β1 and helix α3. Strand β1 hydrogen bonds with and is aligned anti-parallel to strand β5 (where the S2 site is located), and helix α3 packs against both strands. Peptides from these structural elements are highly protected against exchange at early time points, and begin to incorporate additional deuterium over time, indicating that the S2 site is masked, but dynamic enough to incorporate deuterium label over the time course of these experiments. It is likely that interactions of strand β5 with strand β1 and helix α3 play an important role in masking the S2 site, preventing constitutive proteolytic cleavage by ADAM metalloproteases.
Figure 2.
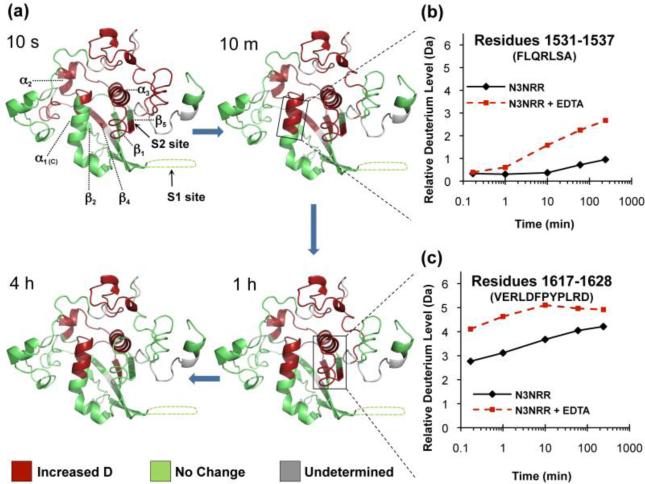
Effect of EDTA treatment on the Notch3 NRR. (a) Time course showing HX-MS of the Notch3 NRR, comparing its closed and open states. The ribbon diagram is colored according to the effect of EDTA treatment on exchange rates: red - regions exhibiting a substantial increase in deuteration upon EDTA treatment (mass difference of 0.8 dalton or more); and green - regions without a major change in deuteration (less than 0.8 dalton change in mass). Areas for which HX-MS data are unavailable are colored in gray. (b) Deuterium incorporation plot of the C-terminal region of helix one (α1) in the presence (red) and absence (black) of EDTA. (c) Deuterium incorporation plot of the peptide that covers the S2 cleavage site in the presence (red) and absence (black) of EDTA. The full dataset for all peptides is in Figure S4B.
The S1 cleavage site is also located in the HD domain, and is predicted to be positioned in an unstructured region (S1 loop) bridging the β3 and β4 strands (Figure 1b, c). Peptides spanning the S1 site undergo rapid exchange, exhibiting maximal uptake (~50% exchange) of deuterium at the first time point, supporting the conclusion that the S1 loop is intrinsically flexible, not strongly hydrogen bonded to other parts of the protein and solvent exposed.
Comparison of the Dynamics of the Notch3 NRR before and after EDTA Treatment
To maintain their structural integrity, the LNR modules of the NRR each rely on three disulfide bonds and a conserved cluster of acidic residues that coordinate a single calcium ion 29. Previous studies have shown that treatment of Notch receptors with divalent cation chelators, such as EDTA, disengages the LNR domain from the interior HD domain, activating Notch signaling, and promoting ectodomain shedding 30,31. We have previously exploited this attribute of the NRR in HX-MS experiments to analyze the extent to which the LNR domain protects the S2 site of human Notch1 22.
Here, we employed a similar approach in order to determine the effect of relaxing the structure of the three LNR modules on the HX-MS profile of the S2 site region of human Notch3, To probe the dynamics of its “open” state, we monitored the pattern of exchange in the EDTA-treated NRR, and then compared it with that of the native, autoinhibited protein (Figure 2a and Figure S4B).
Upon EDTA treatment, the LNR repeats exhibit rapid deuteration, indicating that the protein hydrogen bonding network and or structure is indeed relaxed increasing the deuteration, as predicted once the coordinated ions are removed from the LNRs. In the presence of EDTA, certain regions of the HD domain -- especially at the interdomain interface with the LNR repeats -- also incorporate deuterium more readily. In particular, deuterium exchange is accelerated in peptides encompassing helix α2, which lies near an unstructured loop connecting LNR-B and LNR-C, the top portion of strands β2 and β4, and the C-terminal end of helix α1 (Figure 2a, b). In addition, we observe increased deuterium uptake from the earliest timepoint (10 s) in the peptide spanning the S2 site (Figure 2a, c) and in its proximity, which includes peptides from helix α3 and strand β1. These results confirm the importance of calcium coordination in preserving the overall structure of the NRR by directly maintaining the integrity of the LNR repeats and by indirectly stabilizing the HD domain.
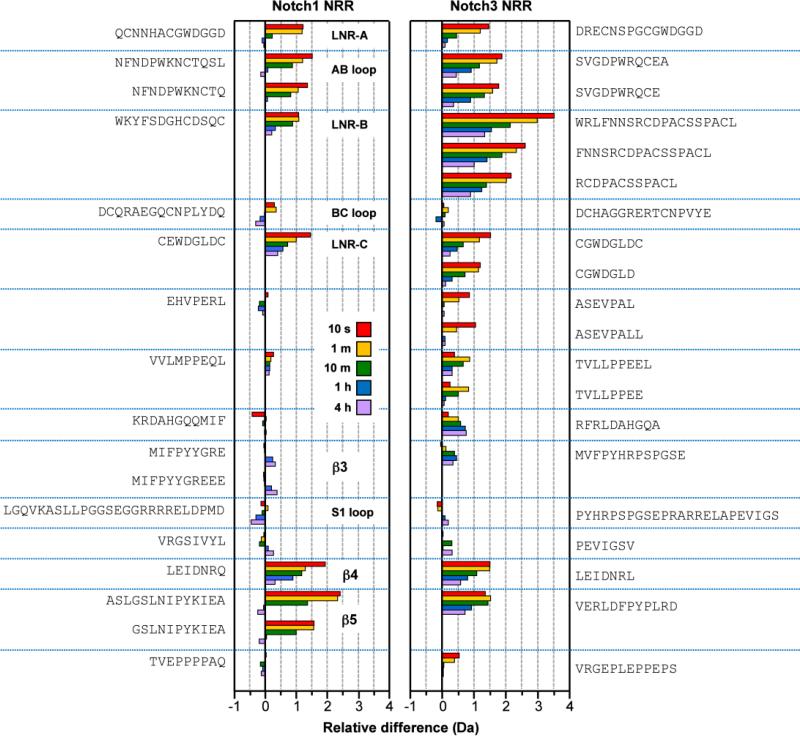
When the exchange properties of the Notch1 and Notch3 NRRs are compared to each other, the most striking difference is in the kinetics of deuteration of LNR-B and the LNR-AB loop. In Notch3, the peptides for these regions exhibit a more dramatic response to EDTA treatment, with an increase in deuteration level of approximately 20%, especially at initial time points. The deuterium uptake patterns between the Notch1 and Notch3 NRRs in other important structural elements are otherwise quite comparable (Figure 3). The increased kinetics of exchange after EDTA treatment, especially in the LNR repeats and parts of the HD domain in contact with the LNRs, reinforce the notion that calcium coordination is critical in maintaining the overall structural integrity of Notch NRRs.
Figure 3.
Comparison of the effect of EDTA treatment on Notch1 and Notch3 NRRs. The plot compares deuterium incorporation levels of Notch1 and Notch3 NRRs as a function of time. Each time point is the relative difference in deuterium incorporation: Notch(EDTA)-Notch(1 or3) and is represented by different color bars. Peptides shown are ordered from N to C terminus (top to bottom). Because the sequences of the Notch1 and Notch3 peptides are not identical, assignments of matched peptides to corresponding structural elements of the proteins are based on sequence alignment, and discretion is advised regarding detailed interpretation.
Molecular Basis of Antibody-Mediated Activation of the Notch3 NRR
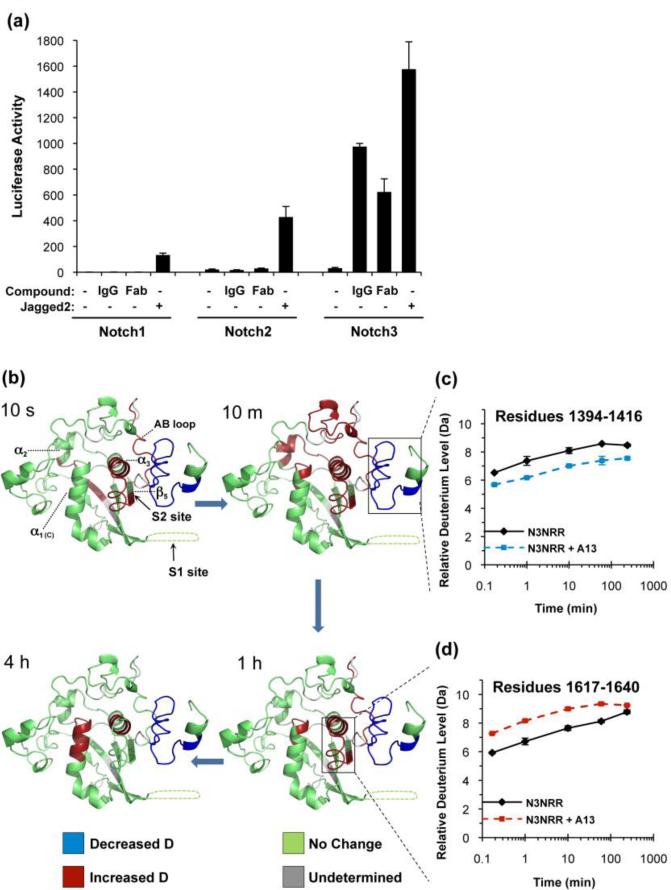
Without ligand engagement, the NRR normally remains in its autoinhibited state and basal Notch signaling activity is minimal. However, the agonist antibody A13, directed against the NRR of human Notch3, can bypass Notch autoinhibition, and activate signaling even in the absence of ligand stimulation 26. Using a luciferase-based reporter assay, we confirmed that the antibody A13 specifically targets the Notch3 NRR, and activates Notch3 in the absence of ligand. The Fab fragment of A13 also stimulates ligand-independent signaling, albeit slightly less effectively than the intact antibody (Figures 4a and Supplementary Figure S5).
Figure 4.
Effects of the A13 antibody on signaling and hydrogen-deuterium exchange of the Notch3 NRR. (a) The A13 antibody specifically activates Notch3 signaling. The effects of both the full IgG and the A13 Fab fragment on Notch activity were tested in co-culture assays. U2OS cells stably expressing Notch-Gal4 chimeric receptors (human Notch1, Notch2 or Notch3) were seeded in 96-well plates pre-coated with normal 3T3 cells or 3T3 cells expressing human Jagged2. The whole IgG and monovalent Fab were used at concentrations of 67 nM and 133 nM, respectively. Firefly luciferase reporter gene activity was measured relative to renilla luciferase as an internal control. Reporter activity from Notch1-Gal4 cells without Jagged2 was arbitrarily set to one; all luciferase measurements are normalized to this control value. All measurements were done in triplicate. Error bars represent standard deviation. (b) The A13 activating antibody binds LNR-A of Notch3 NRR and accelerates deuteration around the S2 site. Ribbon diagram representing the effect of the A13 antibody on the time course of deuterium uptake by the Notch3 NRR. Regions colored blue exhibit a substantial decrease in deuteration in complex with the antibody (mass change of 0.8 dalton or more), whereas regions colored red exhibit a significant increase in deuteration (mass change of 0.8 dalton or more). Regions colored green undergo no major change in deuteration (less than 0.8 dalton change in mass). Regions for which HX-MS data are not available are colored gray. (c) Deuterium incorporation plot of a peptide from LNR-A in the presence and absence of A13 IgG. (d) Deuterium incorporation plot of the peptide that covers the S2 site in the presence and absence of A13 IgG. Error bars represent the range from two data sets. The full dataset for all peptides is shown in Figure S4C.
In order to understand how A13 binding overcomes Notch3 autoinhibition, we compared the HX-MS patterns of the Notch3 NRR in the presence and absence of the A13 antibody. Typically, formation of protein-antibody complexes results in decreased deuterium uptake for peptides stabilized by complex formation, including but not necessarily limited to the antibody epitope 22. In contrast, the complex between A13 and the Notch3 NRR results not only in a retarded rate of deuteration for the peptide spanning the reported epitope, but also in accelerated rates of deuteration for peptides adjacent to the S2 site,
More specifically, upon addition of A13, there is a decrease in deuterium incorporation across all time points for a peptide in the LNR-A module of the Notch3 NRR (Figure 4b, c and Figure S4C). On the other hand, binding of A13 promotes increased deuterium uptake into peptides derived from a linker connecting LNR-A to LNR-B (AB loop), the α3 helix within the HD domain, and a peptide spanning the S2 cleavage site, even at the initial time point (Figure 4b, d). After 10 minutes of deuterium labeling, the elevated deuterium level extends to LNR-B and its contact region of the HD domain at the LNR-HD interface. These results, taken together with previous epitope mapping studies 26, argue that the interaction between A13 and LNR-A sterically interferes with formation of critical interactions between the LNR modules and the HD domain at the interface around the S2 site, thereby trapping the NRR in an “open” state.
Molecular Basis of Antibody-Mediated Inhibition of the Notch3 NRR
Notch antagonists may be of potential therapeutic interest in ovarian cancer and other diseases associated with excess Notch3 signaling 20,21. Several antagonist antibodies have been developed against Notch receptors, which target either the EGF repeats comprising the ligand-binding region, or the NRR. Indeed, antibodies targeting the NRR have been raised against Notch1, Notch2, and Notch3 24-26. All of the anti-NRR antibodies for which epitopes have been mapped bridge the LNR and HD domains, suggesting that they clamp or “lock” the receptor in its quiescent state, but this model has only been probed directly by HX-MS for a single anti-Notch1 antibody, WC-629 24.
To explore whether the clamp model for anti-NRR inhibitory antibodies might apply more broadly, we investigated the mechanism of Notch3 inhibition by the anti-Notch3 NRR antibody A4. After first confirming its inhibitory activity and its specificity for Notch3 (Figure 5), we performed HX-MS on the A4:Notch3-NRR complex in order to compare the exchange patterns of the NRR in the presence and absence of antibody. When the NRR is complexed with A4, we observe decreased deuteration of a peptide in LNR-A, and for several other peptides adjacent to the S1 cleavage site of the HD domain, consistent with the reported26 discontinuous epitope of the antibody (Figure 6a-c and Figure S4D).
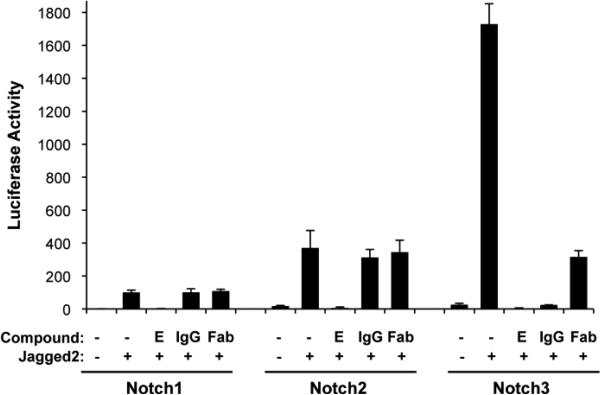
Figure 5.
The A4 antibody specifically inhibits Notch3 signaling. The effects of both the full IgG and the A4 Fab fragment on Notch activity were tested in co-culture assays. U2OS cells stably expressing Notch-Gal4 chimeric receptors (human Notch1, Notch2 or Notch3) were seeded in 96-well plates pre-coated with normal 3T3 cells or 3T3 cells expressing human Jagged2. The whole IgG and the monovalent Fab were used at concentrations of 67 nM and 133 nM, respectively. The gamma secretase inhibitor compound E (E) was used at 1 M as a negative control. Firefly luciferase reporter gene activity was measured relative to renilla luciferase as an internal control. Reporter activity from Notch1-Gal4 cells without Jagged2 was arbitrarily set to one; all luciferase measurements are normalized to this control value. All measurements were done in triplicate. Error bars represent standard deviation.
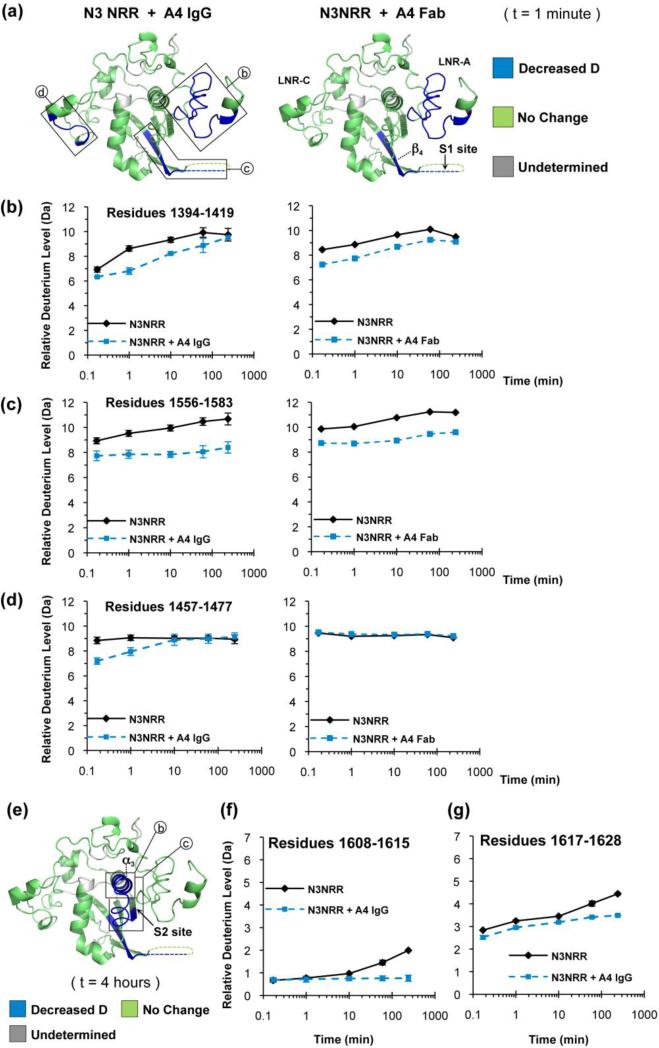
Figure 6.
The A4 inhibitory antibody bridges the LNR and HD domains of the Notch3 NRR. (a) Ribbon diagrams comparing the effects of the A4 IgG (left) and the A4 Fab (right) on the rate of deuterium incorporation into the Notch3 NRR. Regions exhibiting a significant decrease in deuteration in the presence of A4 are colored blue (with a mass change of 0.8 dalton or more). Regions with no major change in deuteration are colored green (with less than 0.8 dalton change in mass). Areas for which HX-MS data are unavailable are colored in gray. The data from the one-minute time point is shown here to illustrate the transient drop in deuteration of a region in LNR-C when complexed with the full A4 IgG, but not with the Fab. (b, c and d) Deuterium incorporation plots of peptides from LNR-A (b), strand β4 and the S1 site (c), and LNR-C (d) when the Notch3 NRR is complexed with the full A4 IgG (left), or the A4 Fab (right). Error bars represent the range from two data sets. The full dataset for all peptides is shown in Figure S2D. (e-g) The A4 inhibitory antibody retards deuteration around the S2 cleavage site. (e) Ribbon diagram representing the effect of the A4 antibody on the extent of deuterium uptake by the Notch3 NRR at the four-hour time point. Peptides showing decreased deuteration in the complex with A4 (a decrease in deuteration level of at least 0.8 dalton) are colored blue, and peptides showing no major change in deuteration (less than 0.8 dalton change in mass) are colored green. (f and g) Deuterium incorporation plots of peptides from helix α3 (f) and from the peptide spanning the S2 site (g) in the presence and absence of the A4 IgG. Error bars represent the range from two data sets.
We also observe slower deuteration kinetics for a peptide located within LNR-C, at a site predicted to be about 40 Å away from the mapped epitope (Figure 6a, d). This region of LNR-C is highly conserved, and overlaps a sequence corresponding to a crystallographic dimer interface seen in the structure of the Notch1 NRR, suggesting that contacts between the two antibody-bound Notch3 NRRs may be induced at this surface upon forming the complex with the full antibody. When we repeated the HX-MS studies using the monovalent Fab fragment of A4 in complex with the Notch3 NRR, deuteration of the LNR-C region in the complex with the Fab was no longer significantly different from the isolated NRR even though the other regions of retarded deuteration were maintained in the complex with the Fab (Figure 6d). Regardless of the origin of the slower deuteration kinetics for this region of LNR-C, its integrity is not required for the antibody to exert its inhibitory effect in cells, as alanine substitutions within this region of LNR-C do not prevent the full antibody from reducing ligand-dependent signals back to basal levels in a reporter gene assay (Figure S6).
Finally, our exchange studies provide direct evidence that binding of the inhibitory antibody reduces the accessibility of the metalloprotease cleavage site of the Notch3 NRR. Upon complexation with antibody, peptides from helix α3 and from the terminal strand housing the S2 cleavage site exhibit reduced deuteration (Figure 6e-g); this effect becomes evident only at later time points for the peptide from helix α3 because it undergoes relatively slow exchange even in the uncomplexed state.
Discussion
Notch signaling participates in numerous cell fate decisions in both development and adult tissue homeostasis. As a result the dose and timing of Notch signals must be precisely regulated. The critical switch controlling ligand responsiveness is the NRR, which remains proteolytically resistant to maintain receptor quiescence at rest, but becomes metalloprotease sensitive after receptor engagement by Delta and Jagged/Serrate family ligands. Further interactions between the ligand-binding, EGF repeats and the NRR may provide an additional layer of receptor regulation, but the extent to which they may contribute to signaling is not clear, and such potential interactions are not addressed in this study.
The studies reported here focus on the NRR of Notch3, because it offers a unique opportunity for investigation of the conformational dynamics of a regulatory domain in a range of different activated and inhibited states by HX-MS. Although antibodies that inhibit signaling of Notch1, Notch2, and Notch3 by binding to the NRR have all been reported24-26, Notch3 is the only receptor for which an activating antibody has also been described 26. Thus, it is possible not only to monitor the conformational dynamics of the Notch3 NRR at rest, in a chemically activated state, and when bound to an inhibitory antibody, but also to uncover how binding of the activating antibody influences the dynamics of the regulatory switch around the metalloprotease cleavage site. Moreover, it is now possible to compare the dynamic profiles of the Notch1 and Notch3 NRRs, because HX-MS studies of the Notch1 NRR in several distinct states have also been previously reported 22.
In these studies, the untreated Notch3 NRR in its autoinhibited conformation shows an overall exchange pattern consistent with the main architectural features of the homology model created based on the X-ray structure of the Notch2 NRR. The most slowly exchanging regions of the protein are located in the HD domain, including the C-terminal end of helix α1, the central region of helix α3 and the internal strands of the beta sheet core. The strand containing the S2 site, along with the C-terminal part of LNR-A, the LNR-AB linker, and LNR-B also show protection from complete exchange at early time points, as predicted for a quiescent, protetolytically resistant state. The Notch3 NRR does appear to be “poised” for release from autoinhibition, however, because the masking of the S2 site is not absolute, it is dynamic and it becomes increasingly deuterated over the four hour experimental time course.
Because LNR modules rely on calcium coordination to maintain their structural integrity 9,29, EDTA treatment has been used to remove bound calcium ions, relax the LNR-HD interface, and thus release autoinhibition of other Notch NRRs 30,31. When Notch3 NRR is treated with EDTA, peptides from the LNR modules exchange rapidly, and regions of the HD domain at the interface with the LNRs also undergo accelerated deuteration by comparison with the untreated NRR. Most notably, peptides flanking the S2 site along with peptides abutting it from helix α3 also show more rapid exchange upon EDTA treatment. These results, taken together with our previous studies showing similar effects in the Notch1 NRR, reinforce and solidify the conclusion that disengagement of the LNR-HD interface is a critical mechanistic step enabling access of the S2 site to metalloproteases. Recently reported studies of the mechanical unfolding of the Notch2 NRR using atomic force microscopy are also consistent with this idea 32.
The most striking findings from these studies come from analysis of the HX-MS patterns of the Notch3 NRR when complexed with the inhibitory and activating antibodies. The eptiopes for these antibodies were precisely mapped when first reported, and the HX-MS data show that peptides encompassing the defined epitopes undergo retarded exchange kinetics upon formation of antibody-NRR complexes. The anti-Notch3 inhibitory antibody has a discontinuous epitope that includes peptides from LNR-A and from the HD domain, as is also seen for the two different Notch1 inhibitory antibodies for which the epitopes are known. The observed protection of a surface on LNR-C upon complexation with the full antibody suggests that the anti-Notch3 inhibitory antibody induces contacts between the two antibody-bound NRR molecules in vitro, because the LNR-C site is far from the primary binding epitope. However, the primary mechanism of antibody-dependent inhibition results from antibody engagement with the primary epitope and does not appear to rely on this region of LNR-C, because alanine substitutions within this site of LNR-C do not affect the capacity of the antibody to reduce ligand-dependent signals back to basal levels in the full-length receptor.
When the NRR is in complex with the activating antibody, the rate of deuteration is retarded for the binding epitope in LNR-A, but accelerated in the adjacent LNR-AB loop, LNR-B, and in the vicinity of the S2 site. Thus, binding of the activating antibody shifts the conformational equilibrium of the adjacent LNR-B domain and otherwise concealed S2 site to the “open” conformation, which is permissive for metalloprotease cleavage. Future studies to determine the ratio of activating antibody affinities for the isolated LNR-A module as compared to the intact Notch3 NRR could be used to estimate the intramolecular conformational equilibrium of the Notch3 NRR for “closed” and “open” states, by analogy to similar studies of the internal equilibrium between active and inactive conformations of tyrosine kinase catalytic domains 33,34.
Given that components of the Notch signaling pathway have been identified as tumor suppressors 35,36 as well as oncogenes, there may be a need for activating antibodies directed against Notch1 and Notch2, in addition to the antibody against Notch3 analyzed here. The mechanistic insights into antibody-mediated activation provided by these studies should facilitate future development of such agents for potential therapeutic exploration.
Materials and Methods
Protein Expression and Purification
The NRRs from human Notch3 (residues S1382 to S1641) and human Notch1 (residues E1446 to Q1733) were subcloned into a pET15b vector containing an N-terminal hexahistidine tag followed by a custom tobacco etch virus (TEV) cleavage site (see Figure S1 for alignment). The purification procedure for each NRR was adapted from previously published methods 9,10,37. Briefly, proteins were produced recombinantly in RosettaII(DE3)pLysS bacteria and recovered from the insoluble fraction using 8M urea. Urea-denatured protein was then affinity-purified on a nickel column, eluted with imidazole and treated with TEV protease to remove the His tag. After TEV treatment, the final form of each NRR contained an additional glycine at its N-terminus. The NRR was then refolded in vitro by dialysis in a redox buffer containing 5mM cysteine and 1mM cystine. The refolded protein was further purified by anion exchange chromatography, followed by size exclusion chromatography in 25 mM Tris buffer pH 8, containing 150 mM NaCl and 10 mM CaCl2. For EDTA experiments, each purified NRR (5 mg/mL) was treated with 20mM EDTA for 2 h at room temperature. The EDTA-treated NRR was directly used in the HX-MS experiment without further purification.
Fab fragment preparation
The Fab fragments of the monoclonal antibodies A13 and A4 were prepared using the Pierce Fab Preparation Kit (Pierce-Thermo, Rockford, IL). Each antibody was digested on a Papain column at 37 °C for 5-6 hours, in a buffer containing 20 mM cysteine at pH7. The digestion mixture was cooled down to ambient temperature, then affinity-purified on a Protein A column in PBS buffer. The eluate was then dialyzed into 25 mM Tris buffer pH 8, containing 150 mM NaCl and 10 mM CaCl2. The Fab fragment from each antibody was further purified by size exclusion chromatography. The efficiency of each digestion and purification step was monitored by SDS-PAGE.
Hydrogen Exchange - Mass Spectrometry Experiments
HX-MS experiments were performed following the previously published report 22 with modifications as described here. A stock solution (150 pmol) of Notch1 NRR or Notch3 NRR, in the presence or absence of EDTA, was prepared in 25 mM Tris buffer pH 8, 150 mM NaCl and 10 mM CaCl2, H2O. For the HX-MS experiment of Notch3 NRR in complex with the whole A13 or A4 antibody, we first incubated 150 pmol of the NRR with 170 pmol of the corresponding antibody for 3 hours in the same buffer. For the experiment monitoring exchange of Notch3 NRR in complex with the A4 Fab fragment, we incubated 150 pmol of the NRR with 300 pmol of the Fab fragment for 3-4 hours in the same buffer. To initiate deuterium exchange, the stock solution was diluted 15-fold with 25mM Tris (p2H 8), 150 mM NaCl and 10 mM CaCl2, 2H2O at 21 °C. At each deuterium exchange time point (10s, 1m, 10m, 1h and 4h), the labeling reaction was stopped by adding an equal volume of quenching buffer (200 mM citrate buffer, 0.5 M TCEP, 4M guanidine HCl, H2O, pH 2.3), and dropping the temperature quickly to 0 °C. In order to reduce the disulfide bonds and allow for efficient pepsin digestion, the reaction was kept at 0 °C for additional 10 minutes in quenching buffer. Each quenched sample was injected into a Waters nanoACQUITY UPLC System with HDX Technology for desalting and subsequent reversed-phase separation of the resulting peptides 38. The separated peptides were directed into a Waters QTof Premier mass spectrophotometer with an electrospray ionization source. The data were analyzed using Masslynx, the excel-based program HX-Express 39 and other custom macros. The data were mapped onto a homology model of human Notch3, which was derived from the Notch2 crystal structure using the Phyre protein fold recognition server 27,28. An absolute difference in mass of 0.8 Da was used as a threshold for all comparisons. This value was well over the measurement error typically obtained from such experimental conditions 40. Any point in the deuterium exchange curves with a difference in mass of less than 0.8 Da was reported as “no change.” The data were not corrected for back exchange and are therefore reported as relative 22.
Luciferase Reporter Assays
Luciferase-based reporter assays were used to assess the efficiency and specificity of antibodies A13 and A4, and their corresponding Fab fragments. In a tissue culture dish (6-cm diameter), U2OS Tet-on flp-in cells bearing isogenic transgenes encoding NOTCH1-, NOTCH2-, or NOTCH3-Gal4 chimeric receptors were transfected with 5 μg of Gal4-firefly luciferase and 100 ng of pRL-TK-Renilla luciferase reporter plasmids. For assays of chimeric Notch3-Gal4 and NRR-Gal4 receptors, U2OS cells were transfected with the indicated Notch3-gal4 construct (1 μg) simultaneously with the reporter plasmids. After 24 hours, cells were split into 96-well plates, which were pre-coated with normal 3T3 or 3T3-Jagged2 cells in the presence of 1 mg/ml tetracycline. The antibody A13 or A4 was added at a final concentration of 67 nM, while Fab fragments were used at 133 nM. The gamma secretase inhibitor, compound E, was used at 1 mM as a negative control. After 24 hours, cells were harvested and lysed in passive lysis buffer. Firefly and renilla luciferase activities in cell lysates were quantitated using a dual luciferase assay (Promega) on a Modulus microplate reader (Turner Biosystems). We also constructed a mutated form of the full-length Notch3 receptor in which four of the residues in the conserved LNR-C patch (E1472 to D1477: EKYCAD, mutated residues in bold) were replaced with alanine in order to test the effect of these mutations on the A4 inhibitory activity in the reporter gene assay.
Supplementary Material
Figure S1. Multiple sequence alignment of the NRR regions of various Notch receptors. The amino acids are colored according to sequence conservation: pink - identical in all sequences; orange - higher than 85% identity or strongly conserved (based on ClustalW); yellow - higher than 55% identity. Secondary structural elements are denoted in white box above the sequences, based on the domain organization of human Notch1NRR. Adapted from reference 9.
Figure S2. Peptide maps of N3NRR. The domain organization of the protein, along with the is defined above the sequence. The color-coded bars below the sequence indicate the peptide fragments that resulted from online pepsin digestion of the Notch3 NRR in various states.
Figure S3. Time course showing the HX-MS behavior of the Notch1 NRR in its “off” state, using the data reported in reference 22. The ribbon diagram is colored according to the relative extent of deuteration of peptides mapped to the indicated regions of the Notch3 NRR, in the same color code used for Figure 1 in the current work (indicated at the bottom of the figure). Sequences for which HX-MS data could not be acquired are colored gray.
Figure S4. Comparison of relative deuterium incorporation for N3NRR in various states. The y axis in each graph represents the relative deuterium level incorporated into each peptide with the maximum indicating the total possible number of exchangeable amide positions. A. The results of three independent experiments of N3NRR alone are presented showing reproducibility. B. Comparison of N3NRR versus N3NRR treated with EDTA. C. Comparison of N3NRR versus N3NRR in complex with A13 IgG. D. Comparison of N3NRR versus N3NRR in complex with A4 IgG. In panels for C and D, plots are shaded blue when peptides show slower deuteration at some point during the time course in the presence of antibody, whereas plots are shaded red when peptides exhibit accelerated deuteration in the presence of antibody.
Figure S5. Intrinsic inhibition of the Notch3 NRR. U2OS cells transiently transfected with the Notch3 NRR were tested for basal activity, activity after A13 treatment, and activity after A4 treatment in luciferase reporter assays (see methods). Firefly luciferase reporter gene activity was measured relative to renilla luciferase as an internal control. Reporter activity from the cells expressing Notch3 NRR without antibody treatment was set to a value of 1; luciferase measurements with antibody treatment were normalized to this control value. All measurements were done in triplicate. Error bars represent standard deviation.
Figure S6. Effect of mutating the LNR-C surface on A4-mediated inhibition of Notch3 signaling. U2OS cells transiently transfected with either wild-type (A) or mutated (B) Notch3 were tested for basal activity, activity upon co-culture with Jagged2-expressing NIH 3T3 cells, and activity upon co-culture with Jagged2-expressing 3T3 cells after A4 antibody treatment using luciferase reporter assays (see methods). Firefly luciferase reporter gene activity was measured relative to renilla luciferase as an internal control. Reporter activity from the cells expressing Notch3 (A) or mutated Notch3 (B) without antibody treatment was set to a relative value of 1; luciferase measurements with co-culture and antibody treatment were normalized to this control value. All measurements were done in triplicate. Error bars represent standard deviation.
Highlights.
Basis for autoinhibition of the Notch3 negative regulatory region and modulation of its activity by allosteric antibodies analyzed using HX-MS
Autoinhibited form of the Notch3 shows retarded deuteration of peptides around the S2 site, showing that key features of autoinhibition are shared among different Notch receptors
Activating antibody traps the Notch3 negative regulatory region in an open state, accelerating deuteration of peptides surrounding the S2 cleavage site
Inhibitory antibody retards deuteration of the S2 site, stabilizing the Notch3 regulatory switch in its autoinhibited state.
Acknowledgements
This work is supported by NIH grants R01 CA092433 (to SCB), NIH awards GM086507 and GM101135 (to JRE), and a research collaboration with the Waters Corporation. We thank Wendy Gordon, Jon Aster, and Roxana Iacob for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am. J. Hum. Genet. 2006;79:169–173. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, Costa T, Pierpont ME, Rand EB, Piccoli DA, Hood L, Spinner NB. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 3.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cécillion M, Marechal E, Maciazek J, Vayssiere C, Cruaud C, Cabanis EA, Ruchoux MM, Weissenbach J, Bach JF, Bousser MG, Tournier-Lasserve E. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 4.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 5.Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Current Opinion in Genetics & Development. 2007;17:52–59. doi: 10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 7.Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, Blacklow SC. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol. 2004;24:9265–9273. doi: 10.1128/MCB.24.21.9265-9273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 10.Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, Blacklow SC. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009;113:4381–4390. doi: 10.1182/blood-2008-08-174748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Israël A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci USA. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 13.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israël A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 14.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 15.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 16.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 17.Petcherski AG, Kimble J. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature. 2000;405:364–368. doi: 10.1038/35012645. [DOI] [PubMed] [Google Scholar]
- 18.Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 19.Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 20.Park JT, Li M, Nakayama K, Mao T-L, Davidson B, Zhang Z, Kurman RJ, Eberhart CG, Shih I-M, Wang T-L. Notch3 gene amplification in ovarian cancer. Cancer Res. 2006;66:6312–6318. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiyanont K, Wales TE, Aste-Amézaga M, Aster JC, Engen JR, Blacklow SC. Evidence for increased exposure of the Notch1 metalloprotease cleavage site upon conversion to an activated conformation. Structure. 2011;19:546–554. doi: 10.1016/j.str.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass spectrometry reviews. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 24.Aste-Amézaga M, Zhang N, Lineberger JE, Arnold BA, Toner TJ, Gu M, Huang L, Vitelli S, Vo KT, Haytko P, Zhao JZ, Baleydier F, L'heureux S, Wang H, Gordon WR, Thoryk E, Andrawes MB, Tiyanont K, Stegmaier K, Roti G, Ross KN, Franlin LL, Wang H, Wang F, Chastain M, Bett AJ, Audoly LP, Aster JC, Blacklow SC, Huber HE. Characterization of notch1 antibodies that inhibit signaling of both normal and mutated notch1 receptors. PLoS ONE. 2010;5:e9094. doi: 10.1371/journal.pone.0009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, Siebel CW. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 26.Li K, Li Y, Wu W, Gordon WR, Chang DW, Lu M, Scoggin S, Fu T, Vien L, Histen G, Zheng J, Martin-Hollister R, Duensing T, Singh S, Blacklow SC, Yao Z, Aster JC, Zhou BBS. Modulation of Notch Signaling by Antibodies Specific for the Extracellular Negative Regulatory Region of NOTCH3. Journal of Biological Chemistry. 2008;283:8046–8054. doi: 10.1074/jbc.M800170200. [DOI] [PubMed] [Google Scholar]
- 27.Bennett-Lovsey RM, Herbert AD, Sternberg MJE, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 28.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 29.Vardar D, North CL, Sanchez-Irizarry C, Aster JC, Blacklow SC. Nuclear magnetic resonance structure of a prototype Lin12-Notch repeat module from human Notch1. Biochemistry. 2003;42:7061–7067. doi: 10.1021/bi034156y. [DOI] [PubMed] [Google Scholar]
- 30.Rand MD, Grimm LM, Artavanis-Tsakonas S, Patriub V, Blacklow SC, Sklar J, Aster JC. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krejcí A, Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephenson NL, Avis JM. Direct observation of proteolytic cleavage at the S2 site upon forced unfolding of the Notch negative regulatory region. Proceedings of the National Academy of Sciences. 2012;109:E2757–65. doi: 10.1073/pnas.1205788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boerner RJ, Kassel DB, Barker SC, Ellis B, DeLacy P, Knight WB. Correlation of the phosphorylation states of pp60c-src with tyrosine kinase activity: the intramolecular pY530-SH2 complex retains significant activity if Y419 is phosphorylated. Biochemistry. 1996;35:9519–9525. doi: 10.1021/bi960248u. [DOI] [PubMed] [Google Scholar]
- 34.LaFevre-Bernt M, Sicheri F, Pico A, Porter M, Kuriyan J, Miller WT. Intramolecular regulatory interactions in the Src family kinase Hck probed by mutagenesis of a conserved tryptophan residue. J Biol Chem. 1998;273:32129–32134. doi: 10.1074/jbc.273.48.32129. [DOI] [PubMed] [Google Scholar]
- 35.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie T-X, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Treviño L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN. Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1. Science. 2011 doi: 10.1126/science.1206923. doi:10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, Leigh IM, Collisson EA, Gordon PB, Jakkula L, Pennypacker S, Zou Y, Sharma M, North JP, Vemula SS, Mauro TM, Neuhaus IM, Leboit PE, Hur JS, Park K, Huh N, Kwok P-Y, Arron ST, Massion PP, Bale AE, Haussler D, Cleaver JE, Gray JW, Spellman PT, South AP, Aster JC, Blacklow SC, Cho RJ. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1114669108. doi:10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordon WR, Vardar-Ulu D, L'heureux S, Ashworth T, Malecki MJ, Sanchez-Irizarry C, Mcarthur DG, Histen G, Mitchell JL, Aster JC, Blacklow SC. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS ONE. 2009;4:e6613. doi: 10.1371/journal.pone.0006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 40.Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci. 2011;100:2071–2086. doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Multiple sequence alignment of the NRR regions of various Notch receptors. The amino acids are colored according to sequence conservation: pink - identical in all sequences; orange - higher than 85% identity or strongly conserved (based on ClustalW); yellow - higher than 55% identity. Secondary structural elements are denoted in white box above the sequences, based on the domain organization of human Notch1NRR. Adapted from reference 9.
Figure S2. Peptide maps of N3NRR. The domain organization of the protein, along with the is defined above the sequence. The color-coded bars below the sequence indicate the peptide fragments that resulted from online pepsin digestion of the Notch3 NRR in various states.
Figure S3. Time course showing the HX-MS behavior of the Notch1 NRR in its “off” state, using the data reported in reference 22. The ribbon diagram is colored according to the relative extent of deuteration of peptides mapped to the indicated regions of the Notch3 NRR, in the same color code used for Figure 1 in the current work (indicated at the bottom of the figure). Sequences for which HX-MS data could not be acquired are colored gray.
Figure S4. Comparison of relative deuterium incorporation for N3NRR in various states. The y axis in each graph represents the relative deuterium level incorporated into each peptide with the maximum indicating the total possible number of exchangeable amide positions. A. The results of three independent experiments of N3NRR alone are presented showing reproducibility. B. Comparison of N3NRR versus N3NRR treated with EDTA. C. Comparison of N3NRR versus N3NRR in complex with A13 IgG. D. Comparison of N3NRR versus N3NRR in complex with A4 IgG. In panels for C and D, plots are shaded blue when peptides show slower deuteration at some point during the time course in the presence of antibody, whereas plots are shaded red when peptides exhibit accelerated deuteration in the presence of antibody.
Figure S5. Intrinsic inhibition of the Notch3 NRR. U2OS cells transiently transfected with the Notch3 NRR were tested for basal activity, activity after A13 treatment, and activity after A4 treatment in luciferase reporter assays (see methods). Firefly luciferase reporter gene activity was measured relative to renilla luciferase as an internal control. Reporter activity from the cells expressing Notch3 NRR without antibody treatment was set to a value of 1; luciferase measurements with antibody treatment were normalized to this control value. All measurements were done in triplicate. Error bars represent standard deviation.
Figure S6. Effect of mutating the LNR-C surface on A4-mediated inhibition of Notch3 signaling. U2OS cells transiently transfected with either wild-type (A) or mutated (B) Notch3 were tested for basal activity, activity upon co-culture with Jagged2-expressing NIH 3T3 cells, and activity upon co-culture with Jagged2-expressing 3T3 cells after A4 antibody treatment using luciferase reporter assays (see methods). Firefly luciferase reporter gene activity was measured relative to renilla luciferase as an internal control. Reporter activity from the cells expressing Notch3 (A) or mutated Notch3 (B) without antibody treatment was set to a relative value of 1; luciferase measurements with co-culture and antibody treatment were normalized to this control value. All measurements were done in triplicate. Error bars represent standard deviation.