Figure 1 (Panels A,B,C).
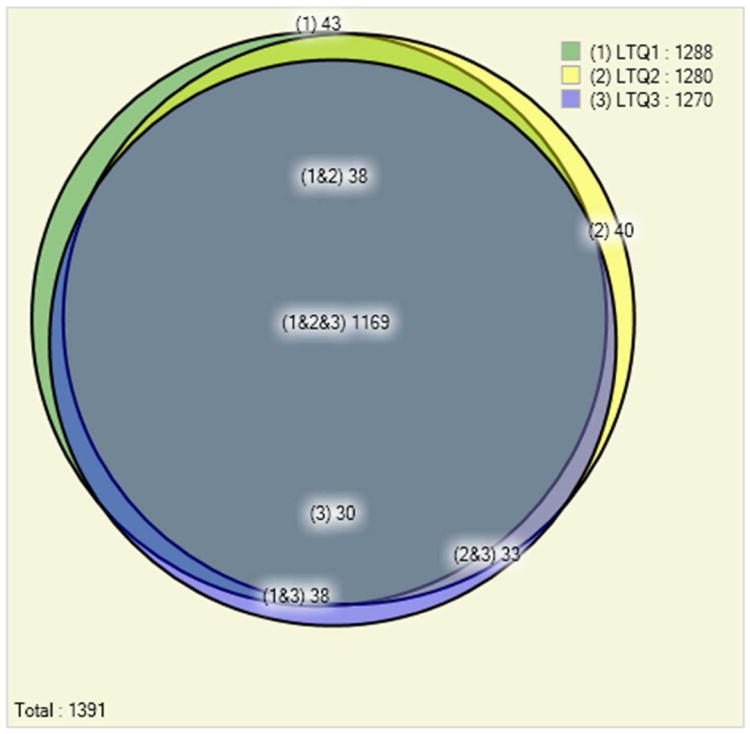
Panel A. Comparison of the number of identified Pyrococcus furiosus proteins in three replicate 12-step MudPIT experiments on an LTQ mass spectrometer at a false discovery rate of below 0.5%. The replicate experiments show high reproducibility of the results: out of the total 1,391 proteins identified 84% (1,169) are identified in each one of the three experiments.
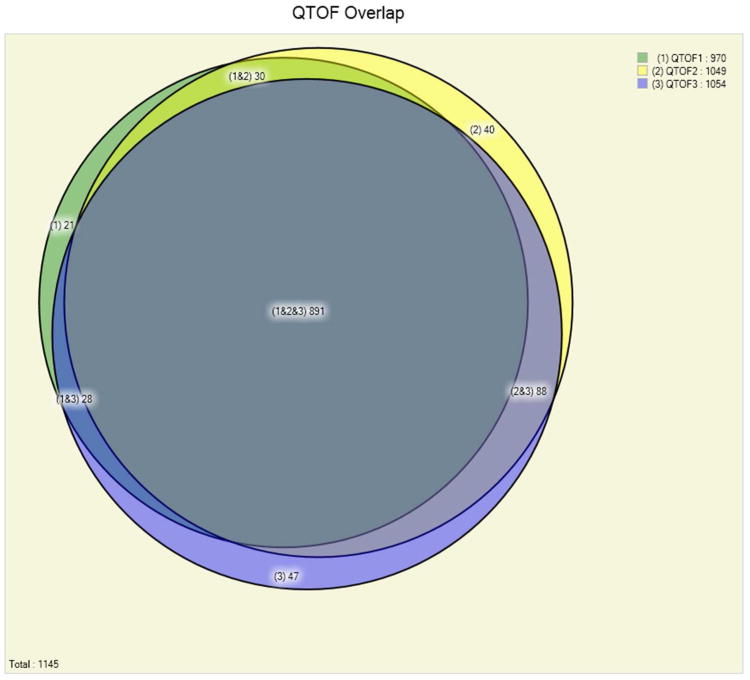
Panel B. Comparison of the number of identified Pyrococcus furiosus proteins in three replicate off-line fractionation experiments on a Q-TOF mass spectrometer at a false discovery rate of below 0.5%. The replicate experiments show high reproducibility of the results: out of the total 1,145 proteins identified 78% (891) are identified in each one of the three experiments.
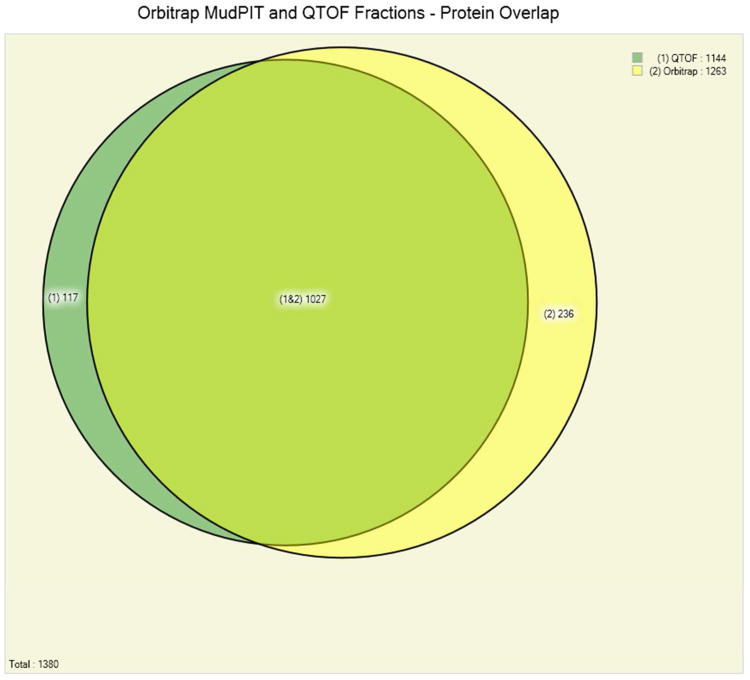
Panel C. Comparison of the overall number of identified Pyrococcus furiosus proteins in 3 protein fractionation experiments on a Q-TOF mass spectrometer and a 12-step MudPIT experiment on an Orbitrap-LTQ mass spectrometer at a false discovery rate of below 0.5%. 1027 of the 1380 identified proteins (74%) are identified by both methods.