Figure 3.
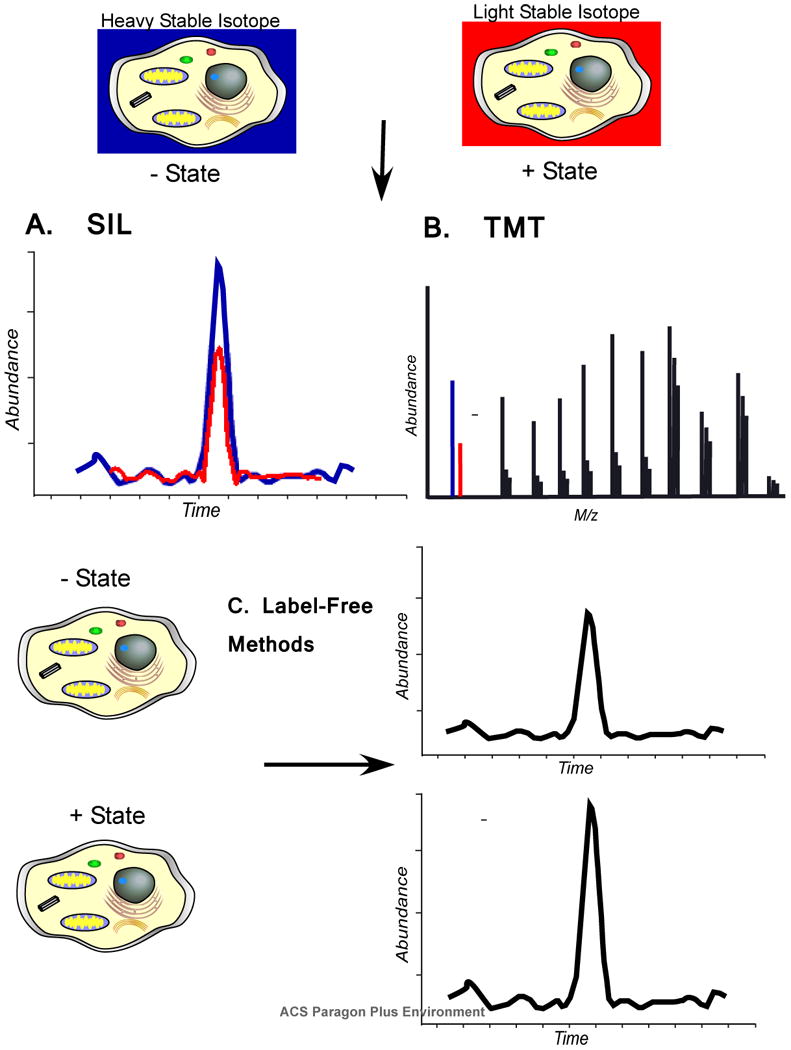
Quantitation of proteins can be performed using stable isotope labels, covalent tags, or label free methods. A) In stable isotope labeling (SIL) methods, a heavy stable isotope label allows peptide masses to be distinguished in the mass spectrometer between different experimental states. Abundance differences can be visualized by selected ion chromatograms. B) Isobaric tags add a mass to the peptides of each state that is isobaric until the peptide ions are fragmented which then reveals a mass difference. The difference in abundance is quantified from the reporter ions in the tandem mass spectrum. C) Two different experimental states can be compared using “label-free” methods. Ion intensity can be measured and compared by selected ion chromatograms as with SIL methods, but these measurements are taken from two different analyses. Another method uses “spectral counting” as a surrogate for abundance based on the observation that proteins which are more abundant have more peptide ions acquired. Label free methods are typically not as accurate as other methods, but can often provide sufficient information to prioritize follow up experiments.
