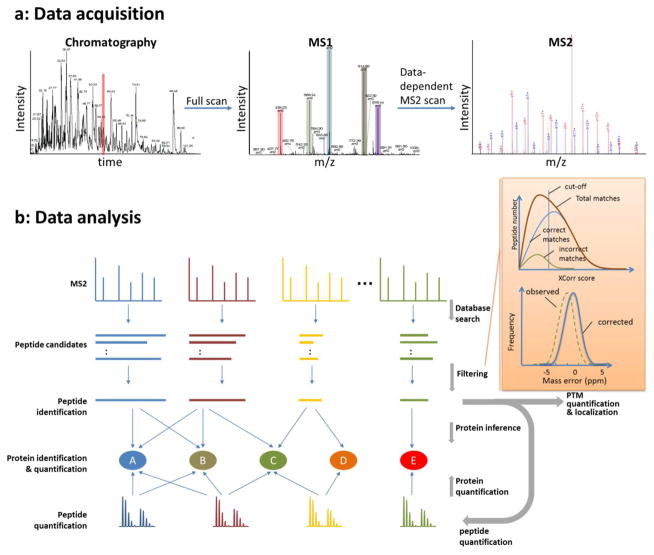
Figure 10.
Representative LC-MS/MS data and a generalized bioinformatic analysis pipeline for protein identification and quantification in shotgun proteomics. (a) As a total ion chromatogram is acquired by nESI-MS from the nLC separation of peptides, the mass spectrometer acquires both full scan MS (MS1) precursor spectra and data-dependent MS/MS (MS2) fragmentation spectra. All ions, typically between 300 – 2000 m/z, are detected during nLC in the full MS scans. The full scan defines the most abundant peptide precursor ions which are sampled by data-dependent for MS/MS. (b) The acquired data is then processed through a bioinformatics pipeline. A database search is used to match theoretically generated peptide fragmentation spectra to experimental MS2 fragmentation spectra, creating a list of peptide candidates for each experimental spectrum. The peptide candidates are ranked and filtered to create peptide spectrum matches (PSMs) and peptide identifications. PSMs can be filtered by XCorr using the “incorrect” reverse PSMs as an estimation of false discovery rate (FDR). In high mass accuracy experiments, prior to XCorr filtering a mass error window can be used to prefilter PSMs based on the precursor mass measurement from MS1 scans. If a systematic mass error is “observed”, it can be “corrected” by adding or subtracting the average mass error. Identified peptides are assigned to proteins by inference to create a list of proteins present within the sample. The relative protein quantification is then performed by averaging the peptide ratio measurements for peptides assigned to the protein. These same strategies can be used for post-translational modification (PTM) identification and quantification, by using a peptide as a surrogate measurement of the PTM.