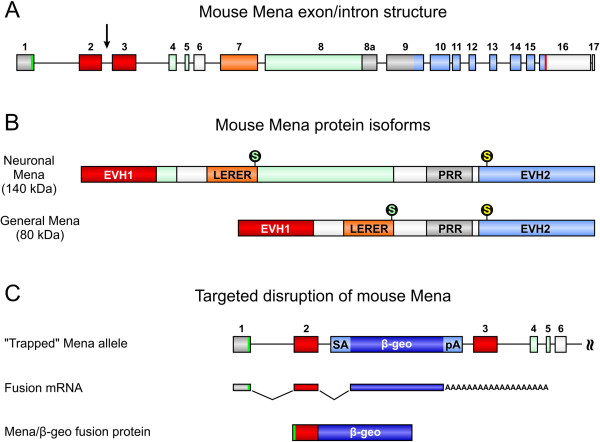
Figure 2.
Targeted disruption of the mouse Mena gene. (A) The mouse Mena gene is located on chromosome 1 spanning about 115 kb of genomic sequence. The translational start and termination codons are given in green (exon 1) and red (exon 16), respectively. The neuronal-specific exons are shown in green. Exons 2 and 3, encoding the Ena/VASP homology 1 (EVH1) domain are shown in red. Exons encoding the LERER repeats, the proline-rich region (PRR), and the EVH2 domain are indicated in orange, grey, and light blue, respectively. The insertion of a gene trap vector in intron 2 of the Mena gene is indicated by a black arrow (compare C). (B) Depending on differential splicing, a neuronal-specific and a general Mena protein isoform exist. Mena is phosphorylated at serine 236 by PKA (green “S”) and at serine 376 by PKG (yellow “S”; numbering according to the 541aa general Mena isoform). (C) In the ES cell clone RRG138, the gene trap vector is inserted in Mena intron 2. The gene trap vector consists of a splice acceptor side (SA), a polyadenylation signal (pA), and β-geo, which encodes β-galactosidase and neomycin resistance [26]. Insertion of the gene trap vector disrupts the Mena gene and results in the expression of a Mena/β-geo fusion protein. Because critical parts of the N-terminal EVH1 domain are missing in the fusion protein, Mena/β-geo most likely lacks all known endogenous Mena functions.