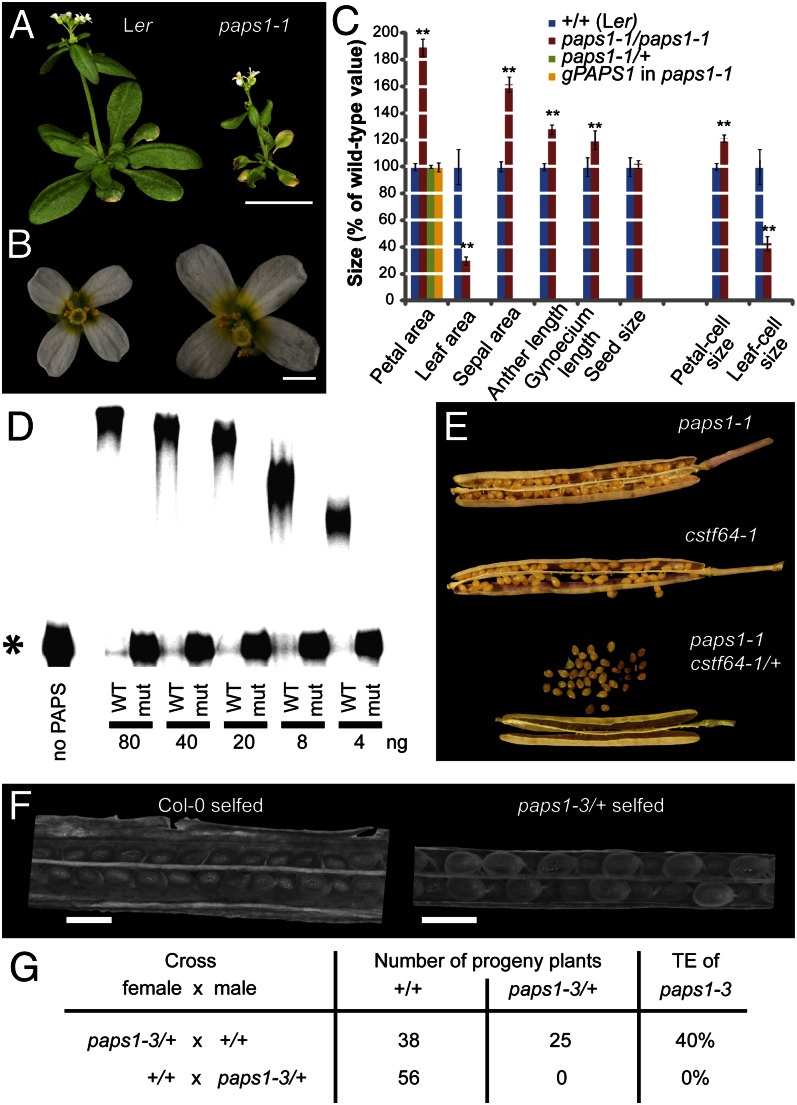
Fig. 1.
Loss of PAPS1 function leads to altered organ growth. (A and B) Whole-plant (A) and flower images (B) of the indicated genotypes. (C) Quantification of organ and cell sizes from the indicated genotypes. Values are mean ± SE from at least 5 (leaves), 20 (petals, sepals, anthers), 7 (gynoecia), or 55 (seeds) organs per genotype, normalized to the wild-type mean. (D) Autoradiograph of in vitro nonspecific polyadenylation assay. The indicated amounts of wild-type PAPS1 protein (WT) or of the mutant form encoded by the paps1-1 allele (mut) were used. Asterisk indicates the unpolyadenylated RNA substrate. (E) Micrographs of opened siliques from the indicated genotypes. Note the aborted seeds produced by paps1-1 cstf64-1/+ plants. (F) Light micrographs of opened siliques from Col-0 (Left) and paps1-3/+ heterozygous plants (Right) after selfing. (G) Transmission efficiency (TE) of the paps1-3 mutant allele through the male and the female gametophyte. The result for the first cross is not significantly different from the expected 50:50 ratio (P = 0.10, χ2 test). (Scale bars, 1 cm in A, 1 mm in B, 500 μm in F.)