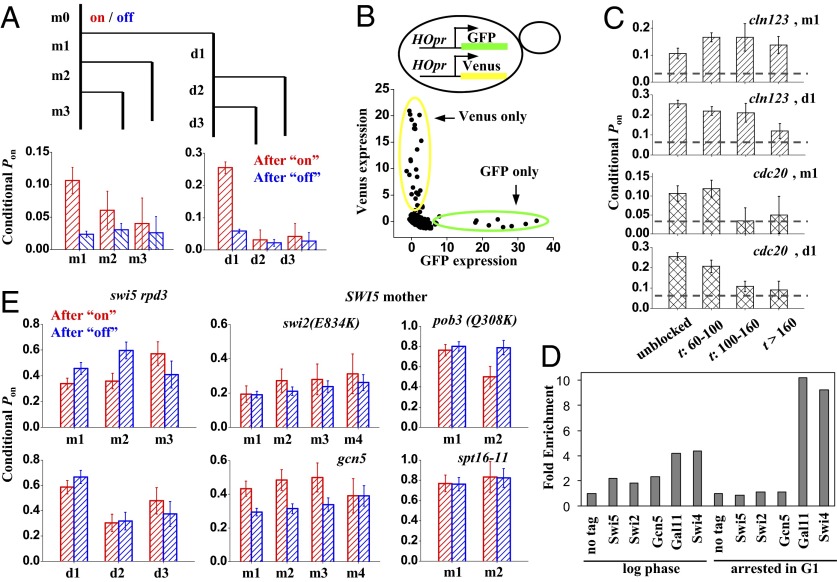
Fig. 3.
Memory of HO expression in the swi5 strain. (A) HO expression in swi5 cells exhibits short-term memory. We divided all cell cycles [mother cycle 0 (m0)] as either “on” (red) or “off” (blue). At the end of m0, the cell divided into mother (m; left branch) and daughter (d; right branch). We followed both cells up to three cycles until the end of the movie (m1–m3 and d1–d3) and recorded the “on/off” status of the HOpr in each of them. Finally, we calculated the Pon in m1–m3 and d1–d3 with either an on or off m0. After an on-cycle, the probability to be on is significantly higher than that after an off-cycle. (B) Coexpression pattern of HOpr-GFP and HOpr-Venus in swi5−/− diploid cells (DY15910). The x axis and y axis for each dot in the plot correspond to the GFP vs. Venus expression level in the same cell cycle (including both mother and daughter). The dots are completely off-diagonal, indicating that the two alleles are activated independently. (C) Memory of HO expression in cell cycles delayed in early G1 (yLB79; cln123, MET-CLN2) or early M (yLB80; cdc20::MET-CDC20). The plots show the Pon in m1 and d1 following a previous on-cycle in either unblocked cells (first bin) or blocked cells with prolonged cell cycle time t (budding-to-budding time). The horizontal lines represent the average Pon, and the existence of memory is indicated by bars significantly higher than this baseline. Memory is quickly lost with a delay in G2/M but not in G1. (D) ChIP experiments show stable Mediator (Gal11) and SBF binding to HOpr following prolonged G1 arrest. Binding of the indicated factors to HOpr was measured by ChIP in growing (log-phase) or arrested cdc28 cells. (E) Memory of HO expression in different genetic backgrounds. In swi5 rpd3 and all SWI5 strains (with the exception of gcn5), the Pon following an on-cycle (red) or an off-cycle (blue) is not significantly different, showing no detectable memory.