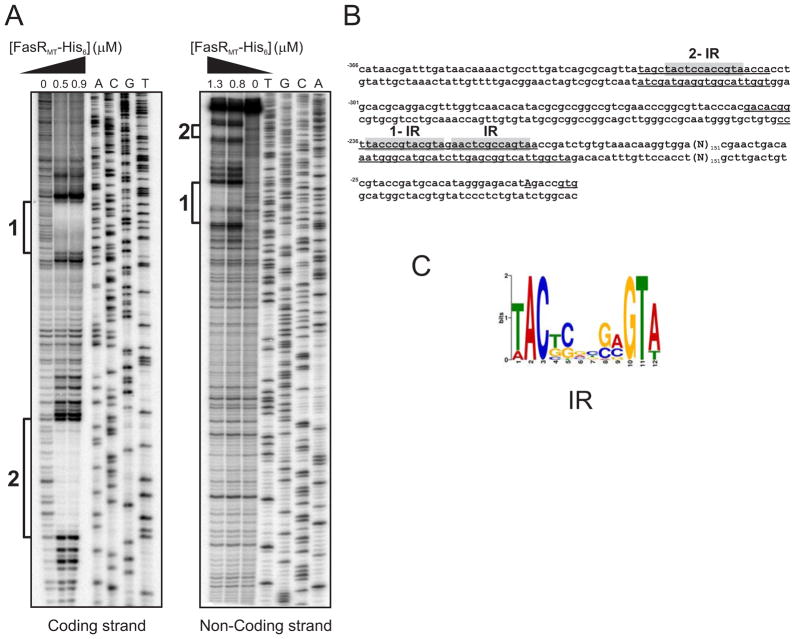
Figure 4. Identification of FasRMT binding sites in the PfasMT promoter region.
A. Both strands containing the PfasMT promoter sequences were labelled with [γ-32P]-ATP and protected from DNase I nuclease activity with two different concentrations of FasRMT (0.5 and 0.9 μM for coding strand; 0.8 and 1.3 μM for non-coding strand). The protected regions in each strand are indicated with black bars. Lanes A–T: DNA sequence of the probe.
B. PfasMT sequence. The protected regions in each strand are underlined. Conserved inverted repeats are highlighted in grey.
C. Sequence analysis of the putative FasR binding regions of several species of mycobacteria with the motif-based sequence analysis tool MEME, led to the identification of a motif highly conserved in Mycobacterium.