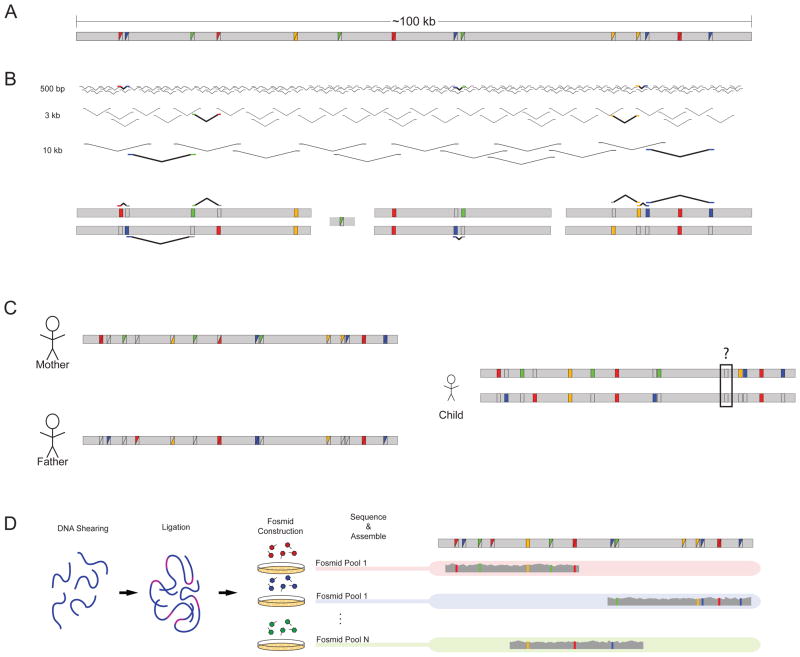
Figure 2. Strategies for empirical haplotype reconstruction.
a | A hypothetical 100 kb stretch of sequence harbours multiple variants compared with the human reference, as designated by the coloured squares. Variants can be homozygous (solid coloured squares) or heterozygous (split coloured squares). b | Sequence reads from libraries of multiple insert sizes can be leveraged to link heterozygous sites together. Informative reads are highlighted and displayed a second time against the diploid reconstruction. The assembly consists of blocks of sequence with gaps arising when variants fall outside the distance of the insert sizes used for sequencing. c | Parental information allows for the separation of chromosomal variants except in instances in which both parents are heterozygous, as demonstrated by the black box in the child’s assembly. d | Laboratory-based methods such as the sequencing of fosmid pools allow for the separation of homologous chromosomes. DNA is sheared, ligated with fosmid vector sequence, packaged and transfected into the bacterium Escherichia coli. Pools of fosmid sequence — each containing only a small fraction of the total genome broken into ~40 kb segments — are sequenced independently. The sequenced libraries are then mapped and assembled for phase reconstruction.