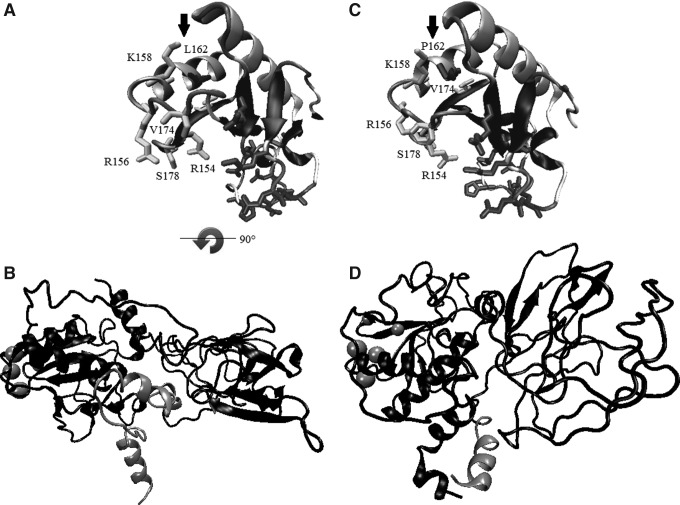
FIG. 3.
Computational approach of the similarities/differences between the wt-DHH (A, B), the p.L162P DHH-N mutant (C), and the Δ1086delG DHH-C mutant (D). In (A, C), the residue in position 162 is represented as bonds and with an arrow. In these panels, only residues in positions 80 to 180 were depicted for clarity. Notable conformational changes were found for the p.L162P mutant protein (C), specifically in the side chains of residues at positions 154, 156, 158, 174, and 178 when compared to the wt-DHH (A). In (B, D) the complete structure of DHH and residues after position 351 are depicted in lighter gray. Residues showed as spheres are those in positions 154, 156, 158, 174, and 178 for spatial reference. The secondary structure for the Δ1086delG DHH-C mutant (D) is modified when compared to the wt-DHH (A).  Indicates a 90° rotation of the (B) figure in relation to the (A) figure.
Indicates a 90° rotation of the (B) figure in relation to the (A) figure.