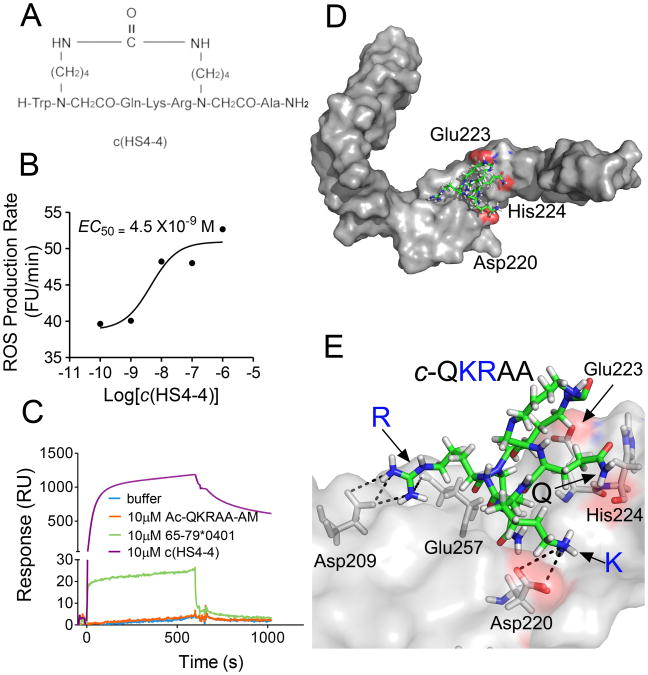
Figure 1.
Chemical structure and ligand properties of c(HS4-4). A. The chemical structure of c(HS4-4), a backbone cyclic peptide analog containing the SE consensus motif QKRAA was designed to induce a stable active conformation. A backbone cyclization methodology was used to keep the functional groups of the side chain residues intact. This feature assures that all the functional groups in a peptide sequence are available for biological activity. B. RAW 264.7 pre-OC cells (3 × 104 /well) were plated in flat-bottom 96-well plates in the presence of various concentrations of c(HS4-4) and ROS production was measured over time. C. SPR data showing binding interactions between recombinant CRT immobilized on a biosensor chip, and c(HS4-4), 15-mer linear SE peptide 65-79*0401, or A 5-mer linear peptide 70-74*0401 (expressing the SE core sequence QKRAA), applied in the analyte. D. A low-power docking image of the c(HS4-4) compound (identified here in its sequence structure cQKRAA, and shown in green) onto the previously identified SE binding site on the CRT P-domain (gray surface). CRT amino acid residues previously found to play critical SE ligand binding roles (7) are highlighted in red. E. A high-power view of c(HS4-4)-CRT molecular interactions. For details see Table 2.