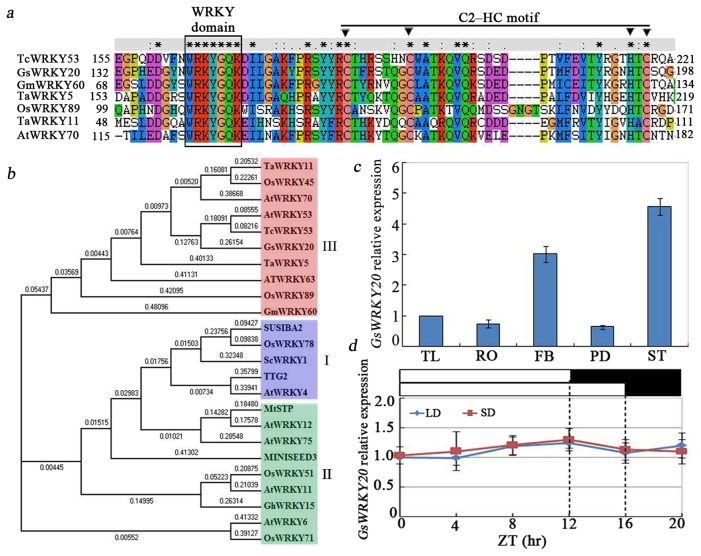
Figure 1. Sequence and expression analysis of GsWRKY20.
(a) Amino acid sequence alignment of WRKY domains among GsWRKY20 and other type Ⅲ WRKY TFs, Sequences were aligned using ClustalW, and gaps were introduced to maximize alignment, filled triangle marks the cystine and histidine in the C2HC-type zinc finger domain.
(b) The phylogenetic tree of the WRKY TFs. The phylogenetic tree was constructed using MEGA 4.1. Total 24 WRKY proteins from Oryza, Arabidopsis, Gossypium, Medicago, Triticum aestivum, Glycine max, Thlaspi caerulescens and Solanum were selected to construct the phylogenetic tree.
(c) Tissue-specific expression analysis of GsWRKY20 by real-time quantitative PCR (qRT-PCR). Tissues included trifoliate leaf (TL), root (RO), stem tip (ST), flower bud (FB), and pod (PD). Expression of GAPDH was used as an internal control. The experiment included three fully independent biological repeats, and three technical repeats and the mean value is shown.
(d) qRT-PCR analysis of GsWRKY20 diurnal expression under SD and LD. Trifoliate leaves were sampled every 4 h at 21 DAE. White and black bars at the top represent light and dark phases, respectively. Relative transcript levels were analyzed by qRT-PCR and normalized by GAPDH. The experiment included three fully independent biological repeats, and three technical repeats and the mean value is shown.