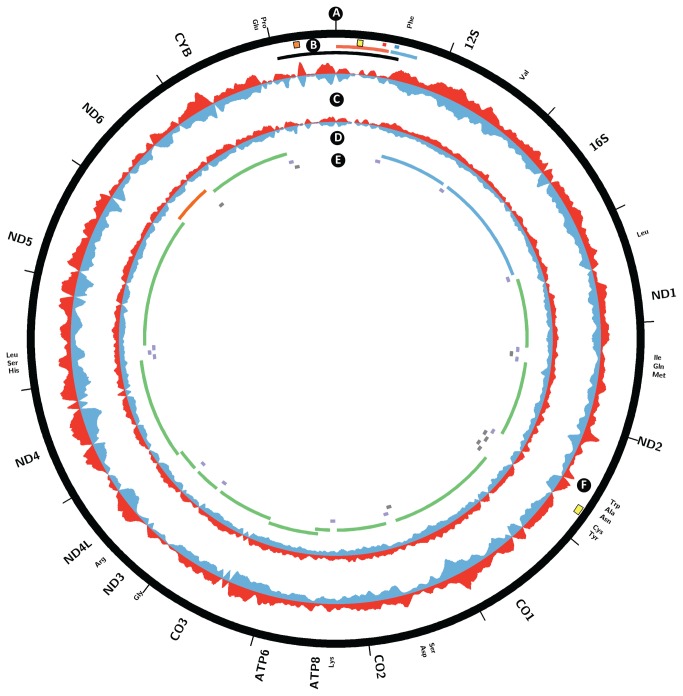
Figure 3. Coating of the mitochondrial genome by TFAM in HeLa cells.
Circos plot of plus strand and minus strand TFAM ChIP-seq and input read density signal over chrM. (A, E) Annotation of protein coding (green on forward/heavy strand, red on reverse/light strand), ribosomal RNA (blue) and tRNA (blue on forward/heavy strand, grey on reverse/light strand) transcripts. (B) D-loop (black), LSP promoter (large red tile), known LSP TFAM binding site (small red tile), HSP promoter (large blue tile), known HSP1 TFAM binding site (small blue tile), and origins of heavy strand replication (Ori-b, orange tile; OH, yellow tile). (C) TFAM ChIP-seq signal on forward (red) and reverse (blue) strands. (D) Input signal on forward (red) and reverse (blue) strands. (F) Origin of light strand replication (yellow tile). Note that the input signal is exaggerated 60-fold relative to the ChIP-seq signal in order to visualize coverage irregularities. The signal from the TFAM ChIP-seq largely follows that of the input, indicating generalized binding across the mitochondrial genome.